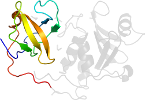Lineage for d2evra2 (2evr A:87-234)
- Root: SCOPe 2.04

Class d: Alpha and beta proteins (a+b) [53931] (380 folds) 
Fold d.3: Cysteine proteinases [54000] (1 superfamily)
consists of one alpha-helix and 4 strands of antiparallel beta-sheet and contains the catalytic triad Cys-His-Asn
Superfamily d.3.1: Cysteine proteinases [54001] (23 families) 
the constitute families differ by insertion into and circular permutation of the common catalytic core made of one alpha-helix and 3-strands of beta-sheet
Family d.3.1.16: NlpC/P60 [142873] (2 proteins)
Pfam PF00877
Protein Cell wall-associated hydrolase Spr C-terminal domain [142874] (1 species) 
Species Nostoc punctiforme [TaxId:272131] [142875] (2 PDB entries)
Uniprot Q8YRR4 87-234
Domain d2evra2: 2evr A:87-234 [132441]
Other proteins in same PDB: d2evra1
complexed with cl, edo, na
Details for d2evra2
PDB Entry: 2evr (more details), 1.6 Å
PDB Description: crystal structure of a putative gamma-d-glutamyl-l-diamino acid endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.60 a resolution
PDB Compounds: (A:) COG0791: Cell wall-associated hydrolases (invasion-associated proteins)SCOPe Domain Sequences for d2evra2:
Sequence; same for both SEQRES and ATOM records: (download)
>d2evra2 d.3.1.16 (A:87-234) Cell wall-associated hydrolase Spr C-terminal domain {Nostoc punctiforme [TaxId: 272131]}
tfseseikkllaeviaftqkamqqsnyylwggtvgpnydcsglmqaafasvgiwlprday
qqegftqpitiaelvagdlvffgtsqkathvglyladgyyihssgkdqgrdgigidilse
qgdavslsyyqqlrgagrvfksyepqrr
SCOPe Domain Coordinates for d2evra2:
Click to download the PDB-style file with coordinates for d2evra2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2evra2:
- d2evra2 first appeared in SCOP 1.73
- d2evra2 appears in SCOPe 2.03
- d2evra2 appears in SCOPe 2.05
- d2evra2 appears in the current release, SCOPe 2.08
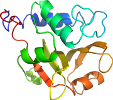
 View in 3D
View in 3D