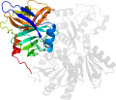
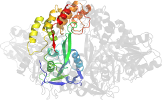
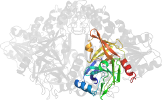
Lineage for d1cu1a3 (1cu1 A:326-631)
- Root: SCOPe 2.07

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.37: P-loop containing nucleoside triphosphate hydrolases [52539] (1 superfamily)
3 layers: a/b/a, parallel or mixed beta-sheets of variable sizes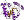
Superfamily c.37.1: P-loop containing nucleoside triphosphate hydrolases [52540] (26 families) 
division into families based on beta-sheet topologies
Family c.37.1.14: RNA helicase [52724] (4 proteins)
duplication: consists of two similar domains, one binds NTP and the other binds RNA; also contains an all-alpha subdomain in the C-terminal extension
Protein HCV helicase domain [52725] (1 species) 
Species Human hepatitis C virus (HCV), different isolates [TaxId:11103] [52726] (17 PDB entries) 
Domain d1cu1a3: 1cu1 A:326-631 [32472]
Other proteins in same PDB: d1cu1a1, d1cu1b1
complexed with po4, zn
Details for d1cu1a3
PDB Entry: 1cu1 (more details), 2.5 Å
PDB Description: crystal structure of an enzyme complex from hepatitis c virus
PDB Compounds: (A:) protein (protease/helicase ns3)SCOPe Domain Sequences for d1cu1a3:
Sequence; same for both SEQRES and ATOM records: (download)
>d1cu1a3 c.37.1.14 (A:326-631) HCV helicase domain {Human hepatitis C virus (HCV), different isolates [TaxId: 11103]}
pgsvtvphpnieevalsntgeipfygkaipieairggrhlifchskkkcdelaaklsglg
inavayyrgldvsviptigdvvvvatdalmtgytgdfdsvidcntcvtqtvdfsldptft
ietttvpqdavsrsqrrgrtgrgrrgiyrfvtpgerpsgmfdssvlcecydagcawyelt
paetsvrlraylntpglpvcqdhlefwesvftglthidahflsqtkqagdnfpylvayqa
tvcaraqapppswdqmwkclirlkptlhgptpllyrlgavqnevtlthpitkyimacmsa
dlevvt
SCOPe Domain Coordinates for d1cu1a3:
Click to download the PDB-style file with coordinates for d1cu1a3.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1cu1a3:

 View in 3D
View in 3D