Lineage for d1zc6a2 (1zc6 A:122-292)
- Root: SCOP 1.75

Class c: Alpha and beta proteins (a/b) [51349] (147 folds) 
Fold c.55: Ribonuclease H-like motif [53066] (7 superfamilies)
3 layers: a/b/a; mixed beta-sheet of 5 strands, order 32145; strand 2 is antiparallel to the rest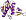
Superfamily c.55.1: Actin-like ATPase domain [53067] (14 families) 
duplication contains two domains of this fold
Family c.55.1.5: BadF/BadG/BcrA/BcrD-like [64089] (5 proteins) 
Protein Probable N-acetylglucosamine kinase CV2896 [142454] (1 species) 
Species Chromobacterium violaceum [TaxId:536] [142455] (1 PDB entry)
Uniprot Q7NU07 122-292! Uniprot Q7NU07 8-121
Domain d1zc6a2: 1zc6 A:122-292 [124889]
Details for d1zc6a2
PDB Entry: 1zc6 (more details), 2.2 Å
PDB Description: Crystal Structure of Putative N-acetylglucosamine Kinase from Chromobacterium violaceum. Northeast Structural Genomics Target Cvr23.
PDB Compounds: (A:) probable N-acetylglucosamine kinaseSCOP Domain Sequences for d1zc6a2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1zc6a2 c.55.1.5 (A:122-292) Probable N-acetylglucosamine kinase CV2896 {Chromobacterium violaceum [TaxId: 536]}
qpgiivalgtgsigealypdgshreaggwgypsgdeasgawlgqraaqltqmaldgrhsh
spltravldfvggdwqammawngratpaqfarlaplvlsaarvdpeadallrqagedawa
iaraldpqdelpvalcgglgqalrdwlppgfrqrlvapqgdsaqgallllq
SCOP Domain Coordinates for d1zc6a2:
Click to download the PDB-style file with coordinates for d1zc6a2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1zc6a2:
- d1zc6a2 first appeared in SCOP 1.73
- d1zc6a2 appears in SCOPe 2.01
- d1zc6a2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D


