Lineage for d1o0qa1 (1o0q A:245-463)
- Root: SCOPe 2.05

Class b: All beta proteins [48724] (176 folds) 
Fold b.80: Single-stranded right-handed beta-helix [51125] (8 superfamilies)
superhelix turns are made of parallel beta-strands and (short) turns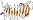
Superfamily b.80.7: beta-Roll [51120] (2 families) 
superhelix turns are made of two short strands each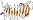
Family b.80.7.1: Serralysin-like metalloprotease, C-terminal domain [51121] (1 protein)
duplication: halfturs of beta-helix are sequence and structural repeats; binds calcium ions between the turns
this is a repeat family; one repeat unit is 1go7 P:374-356 found in domain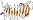
Protein Metalloprotease [51122] (4 species)
The catalytic N-terminal domain belong to the "zincin" superfamily
Species Pseudomonas sp., tac ii 18 [TaxId:306] [82190] (8 PDB entries)
psychrophilic alkaline protease
Domain d1o0qa1: 1o0q A:245-463 [86536]
Other proteins in same PDB: d1o0qa2
complexed with ca, so4
Details for d1o0qa1
PDB Entry: 1o0q (more details), 2.2 Å
PDB Description: Crystal structure of a cold adapted alkaline protease from Pseudomonas TAC II 18, co-crystallized with 1 mM EDTA
PDB Compounds: (A:) serralysinSCOPe Domain Sequences for d1o0qa1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1o0qa1 b.80.7.1 (A:245-463) Metalloprotease {Pseudomonas sp., tac ii 18 [TaxId: 306]}
anletraddtvygfnstadrdfysatsstdklifsvwdgggndtldfsgfsqnqkinlta
gsfsdvggmtgnvsiaqgvtienaiggsgndlligndaanvlkggagndiiyggggadvl
wggtgsdtfvfgavsdstpkaadiikdfqsgfdkidltaitklgglnfvdaftghagdai
vsyhqasnagslqvdfsgqgvadflvttvgqvatydiva
SCOPe Domain Coordinates for d1o0qa1:
Click to download the PDB-style file with coordinates for d1o0qa1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1o0qa1:
- d1o0qa1 first appeared in SCOP 1.65
- d1o0qa1 appears in SCOPe 2.04
- d1o0qa1 appears in SCOPe 2.06
- d1o0qa1 appears in the current release, SCOPe 2.08
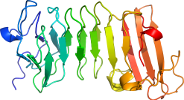
 View in 3D
View in 3D