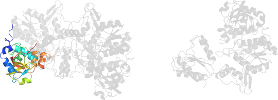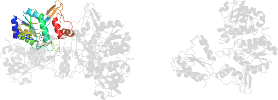Lineage for d1j70c2 (1j70 C:169-389)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.26: Adenine nucleotide alpha hydrolase-like [52373] (3 superfamilies)
core: 3 layers, a/b/a ; parallel beta-sheet of 5 strands, order 32145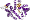
Superfamily c.26.1: Nucleotidylyl transferase [52374] (6 families) 

Family c.26.1.5: ATP sulfurylase catalytic domain [63979] (2 proteins)
automatically mapped to Pfam PF01747
Protein ATP sulfurylase catalytic domain [63980] (5 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [63981] (8 PDB entries) 
Domain d1j70c2: 1j70 C:169-389 [84125]
Other proteins in same PDB: d1j70a1, d1j70a3, d1j70a4, d1j70b1, d1j70b3, d1j70b4, d1j70c1, d1j70c3
complexed with na, po4
Details for d1j70c2
PDB Entry: 1j70 (more details), 2.3 Å
PDB Description: crystal structure of yeast atp sulfurylase
PDB Compounds: (C:) ATP sulphurylaseSCOPe Domain Sequences for d1j70c2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1j70c2 c.26.1.5 (C:169-389) ATP sulfurylase catalytic domain {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
ypglrktpaqlrlefqsrqwdrvvafqtrnpmhrahreltvraareanakvlihpvvglt
kpgdidhhtrvrvyqeiikrypngiaflsllplamrmsgdreavwhaiirknygashfiv
grdhagpgknskgvdfygpydaqelvesykheldievvpfrmvtylpdedryapidqidt
tktrtlnisgtelrrrlrvggeipewfsypevvkilresnp
SCOPe Domain Coordinates for d1j70c2:
Click to download the PDB-style file with coordinates for d1j70c2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1j70c2:
- d1j70c2 first appeared in SCOP 1.65
- d1j70c2 appears in SCOPe 2.07

 View in 3D
View in 3D