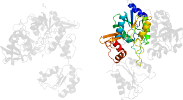Lineage for d1g8ha3 (1g8h A:390-511)
- Root: SCOPe 2.02

Class c: Alpha and beta proteins (a/b) [51349] (147 folds) 
Fold c.37: P-loop containing nucleoside triphosphate hydrolases [52539] (1 superfamily)
3 layers: a/b/a, parallel or mixed beta-sheets of variable sizes
Superfamily c.37.1: P-loop containing nucleoside triphosphate hydrolases [52540] (25 families) 
division into families based on beta-sheet topologies
Family c.37.1.15: ATP sulfurylase C-terminal domain [64011] (1 protein) 
Protein ATP sulfurylase C-terminal domain [64012] (2 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [64013] (7 PDB entries) 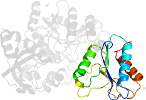
Domain d1g8ha3: 1g8h A:390-511 [60359]
Other proteins in same PDB: d1g8ha1, d1g8ha2, d1g8hb1, d1g8hb2
complexed with acy, adx, ca, cd, mg, na, pop
Details for d1g8ha3
PDB Entry: 1g8h (more details), 2.8 Å
PDB Description: atp sulfurylase from s. cerevisiae: the ternary product complex with aps and ppi
PDB Compounds: (A:) sulfate adenylyltransferaseSCOPe Domain Sequences for d1g8ha3:
Sequence; same for both SEQRES and ATOM records: (download)
>d1g8ha3 c.37.1.15 (A:390-511) ATP sulfurylase C-terminal domain {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
prpkqgfsivlgnsltvsreqlsiallstflqfgggryykifehnnktellsliqdfigs
gsgliipdqweddkdsvvgkqnvylldtsssadiqlesadepishivqkvvlfledngff
vf
SCOPe Domain Coordinates for d1g8ha3:
Click to download the PDB-style file with coordinates for d1g8ha3.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1g8ha3:
- d1g8ha3 first appeared in SCOP 1.57
- d1g8ha3 appears in SCOPe 2.01
- d1g8ha3 appears in SCOPe 2.03
- d1g8ha3 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D