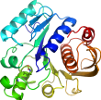
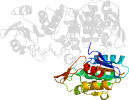
Lineage for d1e8ca3 (1e8c A:104-337)
- Root: SCOPe 2.07

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.72: Ribokinase-like [53612] (3 superfamilies)
core: 3 layers: a/b/a; mixed beta-sheet of 8 strands, order 21345678, strand 7 is antiparallel to the rest
potential superfamily: members of this fold have similar functions but different ATP-binding sites
Superfamily c.72.2: MurD-like peptide ligases, catalytic domain [53623] (3 families) 
has extra strand located between strands 1 and 2
Family c.72.2.1: MurCDEF [53624] (5 proteins)
automatically mapped to Pfam PF08245
Protein UDP-N-acetylmuramyl tripeptide synthetase MurE [64150] (1 species) 
Species Escherichia coli [TaxId:562] [64151] (1 PDB entry) 
Domain d1e8ca3: 1e8c A:104-337 [59379]
Other proteins in same PDB: d1e8ca1, d1e8ca2, d1e8ca4, d1e8cb1, d1e8cb2, d1e8cb4
complexed with api, cl, uag
Details for d1e8ca3
PDB Entry: 1e8c (more details), 2 Å
PDB Description: structure of mure the udp-n-acetylmuramyl tripeptide synthetase from e. coli
PDB Compounds: (A:) udp-n-acetylmuramoylalanyl-d-glutamate--2,6-diaminopimelate ligaseSCOPe Domain Sequences for d1e8ca3:
Sequence; same for both SEQRES and ATOM records: (download)
>d1e8ca3 c.72.2.1 (A:104-337) UDP-N-acetylmuramyl tripeptide synthetase MurE {Escherichia coli [TaxId: 562]}
psdnlrlvgvtgtngkttttqllaqwsqllgeisavmgtvgngllgkviptenttgsavd
vqhelaglvdqgatfcamevsshglvqhrvaalkfaasvftnlsrdhldyhgdmehyeaa
kwllysehhcgqaiinaddevgrrwlaklpdavavsmedhinpnchgrwlkatevnyhds
gatirfssswgdgeieshlmgafnvsnlllalatllalgypladllktaarlqp
SCOPe Domain Coordinates for d1e8ca3:
Click to download the PDB-style file with coordinates for d1e8ca3.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1e8ca3:
- d1e8ca3 first appeared in SCOP 1.57
- d1e8ca3 appears in SCOPe 2.06
- d1e8ca3 appears in the current release, SCOPe 2.08
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d1e8cb1, d1e8cb2, d1e8cb3, d1e8cb4 |