Lineage for Family d.131.1.2: DNA polymerase processivity factor
- Root: SCOPe 2.03

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.131: DNA clamp [55978] (1 superfamily)
contains two helices and two beta sheets
duplication: fold has internal pseudo two-fold symmetry
Superfamily d.131.1: DNA clamp [55979] (3 families) 

Family d.131.1.2: DNA polymerase processivity factor [55983] (5 proteins)
duplication: consists of two domains of this fold
Proteins:
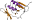
gp45 sliding clamp [55984] (2 species) 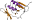
Species Bacteriophage RB69 [TaxId:12353] [55985] (2 PDB entries) 
Species Bacteriophage T4 [TaxId:10665] [55986] (1 PDB entry)

Proliferating cell nuclear antigen (PCNA) [55989] (6 species) 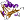
Species Archaeoglobus fulgidus [TaxId:2234] [103254] (3 PDB entries) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [55990] (5 PDB entries)
Uniprot P15873
Species Haloferax volcanii [TaxId:2246] [225733] (1 PDB entry) 
Species Human (Homo sapiens) [TaxId:9606] [55991] (10 PDB entries)
Uniprot P12004
Species Pyrococcus furiosus [TaxId:2261] [55992] (4 PDB entries) 
Species Sulfolobus tokodaii [TaxId:111955] [111175] (1 PDB entry)
Uniprot Q975N2

UL42 [55987] (1 species) 
Species Human herpesvirus type 1 [TaxId:10298] [55988] (1 PDB entry)
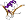
UL44 [111176] (1 species) 
Species Human cytomegalovirus, HHV-5 [TaxId:10359] [111177] (2 PDB entries)
Uniprot P16790 1-290

automated matches [227006] (3 species)
not a true protein
Species Pyrococcus furiosus [TaxId:2261] [225778] (1 PDB entry) 
Species Saccharomyces cerevisiae [TaxId:4932] [232312] (4 PDB entries) 
Species Saccharomyces cerevisiae [TaxId:559292] [233826] (2 PDB entries)
More info for Family d.131.1.2: DNA polymerase processivity factor
Timeline for Family d.131.1.2: DNA polymerase processivity factor:
- Family d.131.1.2: DNA polymerase processivity factor first appeared (with stable ids) in SCOP 1.55
- Family d.131.1.2: DNA polymerase processivity factor appears in SCOPe 2.02
- Family d.131.1.2: DNA polymerase processivity factor appears in SCOPe 2.04
- Family d.131.1.2: DNA polymerase processivity factor appears in the current release, SCOPe 2.08