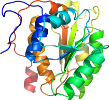
Lineage for d7u1da1 (7u1d A:86-280)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.36: Thiamin diphosphate-binding fold (THDP-binding) [52517] (1 superfamily)
3 layers: a/b/a; parallel beta-sheet of 6 strands, order 213465
Superfamily c.36.1: Thiamin diphosphate-binding fold (THDP-binding) [52518] (9 families) 
there are two different functional modules of this fold: pyridine-binding (Pyr) and pyrophosphate-binding (PP) modules
two Pyr and two PP modules assemble together in a conserved heterotetrameric core that binds two THDP coenzyme molecules
Family c.36.1.0: automated matches [227300] (1 protein)
not a true family
Protein automated matches [227126] (21 species)
not a true protein
Species Thale cress (Arabidopsis thaliana) [TaxId:3702] [255810] (3 PDB entries) 
Domain d7u1da1: 7u1d A:86-280 [422881]
Other proteins in same PDB: d7u1da2
automated match to d1ybha2
complexed with cie, f50, fad, mg, nhe, so4, tp9; mutant
Details for d7u1da1
PDB Entry: 7u1d (more details), 3.11 Å
PDB Description: crystal structure of arabidopsis thaliana acetohydroxyacid synthase p197t mutant in complex with chlorimuron-ethyl
PDB Compounds: (A:) Acetolactate synthase, chloroplasticSCOPe Domain Sequences for d7u1da1:
Sequence; same for both SEQRES and ATOM records: (download)
>d7u1da1 c.36.1.0 (A:86-280) automated matches {Thale cress (Arabidopsis thaliana) [TaxId: 3702]}
tfisrfapdqprkgadilvealerqgvetvfaypggasmeihqaltrsssirnvlprheq
ggvfaaegyarssgkpgiciatsgpgatnlvsgladalldsvplvaitgqvtrrmigtda
fqetpivevtrsitkhnylvmdvedipriieeafflatsgrpgpvlvdvpkdiqqqlaip
nweqamrlpgymsrm
SCOPe Domain Coordinates for d7u1da1:
Click to download the PDB-style file with coordinates for d7u1da1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d7u1da1:
- d7u1da1 is new in SCOPe 2.08-stable
 View in 3D
View in 3D
