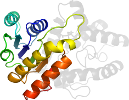
Lineage for d1emd_2 (1emd 146-312)
- Root: SCOP 1.67

Class d: Alpha and beta proteins (a+b) [53931] (260 folds) 
Fold d.162: LDH C-terminal domain-like [56326] (1 superfamily)
unusual fold, defines family
Superfamily d.162.1: LDH C-terminal domain-like [56327] (2 families) 
Family d.162.1.1: Lactate & malate dehydrogenases, C-terminal domain [56328] (4 proteins)
N-terminal domain is NAD-binding module (alpha/beta Rossmann-fold domain)
Protein Malate dehydrogenase [56329] (11 species) 
Species Escherichia coli [TaxId:562] [56333] (4 PDB entries) 
Domain d1emd_2: 1emd 146-312 [42103]
Other proteins in same PDB: d1emd_1
complexed with cit, nad
Details for d1emd_2
PDB Entry: 1emd (more details), 1.9 Å
PDB Description: crystal structure of a ternary complex of escherichia coli malate dehydrogenase, citrate and nad at 1.9 angstroms resolution
SCOP Domain Sequences for d1emd_2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1emd_2 d.162.1.1 (146-312) Malate dehydrogenase {Escherichia coli}
vttldiirsntfvaelkgkqpgevevpvigghsgvtilpllsqvpgvsfteqevadltkr
iqnagtevveakagggsatlsmgqaaarfglslvralqgeqgvvecayvegdgqyarffs
qplllgkngveerksigtlsafeqnalegmldtlkkdialgqefvnk
SCOP Domain Coordinates for d1emd_2:
Click to download the PDB-style file with coordinates for d1emd_2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1emd_2:
- d1emd_2 first appeared (with stable ids) in SCOP 1.55
- d1emd_2 appears in SCOP 1.65
- d1emd_2 appears in SCOP 1.69
- d1emd_2 appears in the current release, SCOPe 2.08, called d1emda2
 View in 3D
View in 3D