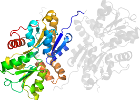Lineage for d7ahrb1 (7ahr B:2-282)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.94: Periplasmic binding protein-like II [53849] (1 superfamily)
consists of two similar intertwined domain with 3 layers (a/b/a) each: duplication
mixed beta-sheet of 5 strands, order 21354; strand 5 is antiparallel to the rest
Superfamily c.94.1: Periplasmic binding protein-like II [53850] (4 families) 
Similar in architecture to the superfamily I but partly differs in topology
Family c.94.1.1: Phosphate binding protein-like [53851] (45 proteins)
has additional insertions and/or extensions that are not grouped together

Protein automated matches [190140] (37 species)
not a true protein
Species Streptomyces coelicolor [TaxId:100226] [420521] (1 PDB entry) 
Domain d7ahrb1: 7ahr B:2-282 [420562]
Other proteins in same PDB: d7ahra2, d7ahra3, d7ahrb2, d7ahrb3
automated match to d2nxoa1
complexed with gol, rdh
Details for d7ahrb1
PDB Entry: 7ahr (more details), 2.21 Å
PDB Description: enzyme of biosynthetic pathway
PDB Compounds: (B:) Chorismate dehydrataseSCOPe Domain Sequences for d7ahrb1:
Sequence; same for both SEQRES and ATOM records: (download)
>d7ahrb1 c.94.1.1 (B:2-282) automated matches {Streptomyces coelicolor [TaxId: 100226]}
dnsrtrprvghiqflnclplywglartgtlldfeltkdtpeklseqlvrgdldigpvtlv
eflknaddlvafpdiavgcdgpvmscvivsqvpldrldgarvalgstsrtsvrlaqllls
erfgvqpdyytcppdlslmmqeadaavligdaalranmidgprygldvhdlgalwkewtg
lpfvfavwaarrdyaerepvitrkvheaflasrnlsleevekvaeqaarweafdedtlak
yfttldfrfgapqleavtefarrvgpttgfpadvkvellkp
SCOPe Domain Coordinates for d7ahrb1:
Click to download the PDB-style file with coordinates for d7ahrb1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d7ahrb1:
- d7ahrb1 is new in SCOPe 2.08-stable

 View in 3D
View in 3D