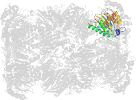
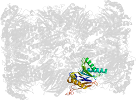
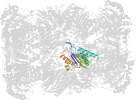
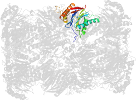
Lineage for d1g0uk_ (1g0u K:)
- Root: SCOP 1.57

Class d: Alpha and beta proteins (a+b) [53931] (194 folds) 
Fold d.153: N-terminal nucleophile aminohydrolases (Ntn hydrolases) [56234] (1 superfamily) 
Superfamily d.153.1: N-terminal nucleophile aminohydrolases (Ntn hydrolases) [56235] (5 families) 

Family d.153.1.4: Proteasome subunits [56251] (3 proteins) 
Protein Proteasome beta subunit (catalytic) [56252] (2 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [56254] (3 PDB entries) 
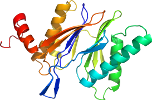
Domain d1g0uk_: 1g0u K: [41893]
Other proteins in same PDB: d1g0ua_, d1g0ub_, d1g0uc_, d1g0ud_, d1g0ue_, d1g0uf_, d1g0ug_, d1g0uo_, d1g0up_, d1g0uq_, d1g0ur_, d1g0us_, d1g0ut_, d1g0uu_
Details for d1g0uk_
PDB Entry: 1g0u (more details), 2.4 Å
PDB Description: a gated channel into the proteasome core particle
SCOP Domain Sequences for d1g0uk_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1g0uk_ d.153.1.4 (K:) Proteasome beta subunit (catalytic) {Baker's yeast (Saccharomyces cerevisiae)}
tttlafrfqggiivavdsratagnwvasqtvkkvieinpfllgtmaggaadcqfwetwlg
sqcrlhelrekerisvaaaskilsnlvyqykgaglsmgtmicgytrkegptiyyvdsdgt
rlkgdifcvgsgqtfaygvldsnykwdlsvedalylgkrsilaaahrdaysggsvnlyhv
tedgwiyhgnhdvgelfwkvkeeegsfnnvig
SCOP Domain Coordinates for d1g0uk_:
Click to download the PDB-style file with coordinates for d1g0uk_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1g0uk_:
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d1g0u1_, d1g0u2_, d1g0ua_, d1g0ub_, d1g0uc_, d1g0ud_, d1g0ue_, d1g0uf_, d1g0ug_, d1g0uh_, d1g0ui_, d1g0uj_, d1g0ul_, d1g0um_, d1g0un_, d1g0uo_, d1g0up_, d1g0uq_, d1g0ur_, d1g0us_, d1g0ut_, d1g0uu_, d1g0uv_, d1g0uw_, d1g0ux_, d1g0uy_, d1g0uz_ |