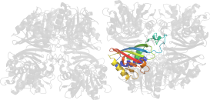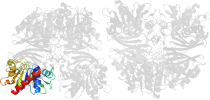Lineage for d1cerr2 (1cer R:149-312)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.81: FwdE/GAPDH domain-like [55346] (4 superfamilies)
core: alpha-beta-alpha-beta(3); mixed sheet: 2134, strand 2 is parallel to strand 1
Superfamily d.81.1: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain [55347] (5 families) 
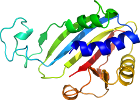
N-terminal domain is the classic Rossmann-fold
Family d.81.1.1: GAPDH-like [55348] (6 proteins)
has many additional secondary structures
Protein Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [55349] (21 species) 
Species Thermus aquaticus [TaxId:271] [55352] (2 PDB entries) 
Domain d1cerr2: 1cer R:149-312 [39904]
Other proteins in same PDB: d1cera1, d1cerb1, d1cerc1, d1cerd1, d1cero1, d1cerp1, d1cerq1, d1cerr1
complexed with nad
Details for d1cerr2
PDB Entry: 1cer (more details), 2.5 Å
PDB Description: determinants of enzyme thermostability observed in the molecular structure of thermus aquaticus d-glyceraldehyde-3-phosphate dehydrogenase at 2.5 angstroms resolution
PDB Compounds: (R:) holo-d-glyceraldehyde-3-phosphate dehydrogenaseSCOPe Domain Sequences for d1cerr2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1cerr2 d.81.1.1 (R:149-312) Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) {Thermus aquaticus [TaxId: 271]}
cttnslapvmkvleeafgvekalmttvhsytndqrlldlphkdlrraraaainiiptttg
aakatalvlpslkgrfdgmalrvptatgsisditallkrevtaeevnaalkaaaegplkg
ilaytedeivlqdivmdphssivdakltkalgnmvkvfawyd
SCOPe Domain Coordinates for d1cerr2:
Click to download the PDB-style file with coordinates for d1cerr2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1cerr2:
- d1cerr2 first appeared (with stable ids) in SCOP 1.55
- d1cerr2 appears in SCOPe 2.07
 View in 3D
View in 3D