Lineage for d4qz7q1 (4qz7 Q:1-234)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.153: Ntn hydrolase-like [56234] (2 superfamilies)
4 layers: alpha/beta/beta/alpha; has an unusual sheet-to-sheet packing
Superfamily d.153.1: N-terminal nucleophile aminohydrolases (Ntn hydrolases) [56235] (8 families) 
N-terminal residue provides two catalytic groups, nucleophile and proton donor
Family d.153.1.0: automated matches [191393] (1 protein)
not a true family
Protein automated matches [190509] (19 species)
not a true protein
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:559292] [256123] (110 PDB entries) 
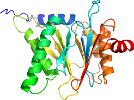
Domain d4qz7q1: 4qz7 Q:1-234 [309236]
Other proteins in same PDB: d4qz7a_, d4qz7b_, d4qz7c2, d4qz7d_, d4qz7e_, d4qz7f_, d4qz7g_, d4qz7h_, d4qz7i_, d4qz7j_, d4qz7k_, d4qz7l_, d4qz7m_, d4qz7n_, d4qz7o_, d4qz7p_, d4qz7q2, d4qz7r_, d4qz7s_, d4qz7t_, d4qz7u_, d4qz7v_, d4qz7w_, d4qz7x_, d4qz7y_, d4qz7z_
automated match to d4eu2a_
complexed with 04c, cl, mes, mg; mutant
Details for d4qz7q1
PDB Entry: 4qz7 (more details), 2.8 Å
PDB Description: yCP beta5-A50V mutant in complex with the epoxyketone inhibitor ONX 0914
PDB Compounds: (Q:) Proteasome subunit alpha type-4SCOPe Domain Sequences for d4qz7q1:
Sequence; same for both SEQRES and ATOM records: (download)
>d4qz7q1 d.153.1.0 (Q:1-234) automated matches {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 559292]}
gydralsifspdghifqveyaleavkrgtcavgvkgkncvvlgcerrstlklqdtritps
kvskidshvvlsfsglnadsriliekarveaqshrltledpvtveyltryvagvqqrytq
sggvrpfgvstliagfdprddepklyqtepsgiysswsaqtigrnsktvrefleknydrk
eppatveecvkltvrsllevvqtgaknieitvvkpdsdivalsseeinqyvtqi
SCOPe Domain Coordinates for d4qz7q1:
Click to download the PDB-style file with coordinates for d4qz7q1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4qz7q1:
- d4qz7q1 first appeared in SCOPe 2.06, called d4qz7q_
- d4qz7q1 was called d4qz7q_ in SCOPe 2.07
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d4qz7a_, d4qz7b_, d4qz7c1, d4qz7c2, d4qz7d_, d4qz7e_, d4qz7f_, d4qz7g_, d4qz7h_, d4qz7i_, d4qz7j_, d4qz7k_, d4qz7l_, d4qz7m_, d4qz7n_, d4qz7o_, d4qz7p_, d4qz7r_, d4qz7s_, d4qz7t_, d4qz7u_, d4qz7v_, d4qz7w_, d4qz7x_, d4qz7y_, d4qz7z_ |