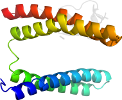
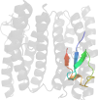Lineage for d3pzab2 (3pza B:135-171)
- Root: SCOPe 2.04

Class g: Small proteins [56992] (91 folds) 
Fold g.41: Rubredoxin-like [57769] (17 superfamilies)
metal(zinc or iron)-bound fold; sequence contains two CX(n)C motifs, in most cases n = 2
Superfamily g.41.5: Rubredoxin-like [57802] (4 families) 

Family g.41.5.0: automated matches [232942] (1 protein)
not a true family
Protein automated matches [232943] (2 species)
not a true protein
Species Pyrococcus furiosus [TaxId:2261] [232944] (4 PDB entries) 
Domain d3pzab2: 3pza B:135-171 [248734]
Other proteins in same PDB: d3pzaa1, d3pzab1
automated match to d3mpsa2
complexed with fe2, peo
Details for d3pzab2
PDB Entry: 3pza (more details), 1.85 Å
PDB Description: Fully Reduced (All-ferrous) Pyrococcus rubrerythrin after a 10 second exposure to peroxide.
PDB Compounds: (B:) rubrerythrinSCOPe Domain Sequences for d3pzab2:
Sequence; same for both SEQRES and ATOM records: (download)
>d3pzab2 g.41.5.0 (B:135-171) automated matches {Pyrococcus furiosus [TaxId: 2261]}
eikkvyicpicgytavdeapeycpvcgapkekfvvfe
SCOPe Domain Coordinates for d3pzab2:
Click to download the PDB-style file with coordinates for d3pzab2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3pzab2:
- d3pzab2 is new in SCOPe 2.04-stable
- d3pzab2 appears in SCOPe 2.05
- d3pzab2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D