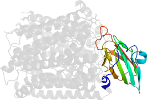Lineage for d1ehkb1 (1ehk B:41-168)
- Root: SCOP 1.69

Class b: All beta proteins [48724] (144 folds) 
Fold b.6: Cupredoxin-like [49502] (2 superfamilies)
sandwich; 7 strands in 2 sheets, greek-key
variations: some members have additional 1-2 strands
Superfamily b.6.1: Cupredoxins [49503] (6 families) 
contains copper-binding site
Family b.6.1.2: Periplasmic domain of cytochrome c oxidase subunit II [49541] (2 proteins) 
Protein Cytochrome c oxidase [49544] (4 species) 
Species Thermus thermophilus, ba3 type [TaxId:274] [49547] (2 PDB entries) 
Domain d1ehkb1: 1ehk B:41-168 [23041]
Other proteins in same PDB: d1ehka_, d1ehkb2, d1ehkc_
Details for d1ehkb1
PDB Entry: 1ehk (more details), 2.4 Å
PDB Description: crystal structure of the aberrant ba3-cytochrome-c oxidase from thermus thermophilus
SCOP Domain Sequences for d1ehkb1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1ehkb1 b.6.1.2 (B:41-168) Cytochrome c oxidase {Thermus thermophilus, ba3 type}
tagvipagklervdpttvrqegpwadpaqavvqtgpnqytvyvlafafgyqpnpievpqg
aeivfkitspdvihgfhvegtninvevlpgevstvrytfkrpgeyriicnqycglghqnm
fgtivvke
SCOP Domain Coordinates for d1ehkb1:
Click to download the PDB-style file with coordinates for d1ehkb1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1ehkb1:
- d1ehkb1 first appeared (with stable ids) in SCOP 1.55
- d1ehkb1 appears in SCOP 1.67
- d1ehkb1 appears in SCOP 1.71
- d1ehkb1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D