Lineage for d4hcla2 (4hcl A:127-382)
- Root: SCOPe 2.08

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.1: TIM beta/alpha-barrel [51350] (34 superfamilies)
contains parallel beta-sheet barrel, closed; n=8, S=8; strand order 12345678
the first seven superfamilies have similar phosphate-binding sites
Superfamily c.1.11: Enolase C-terminal domain-like [51604] (3 families) 
binds metal ion (magnesium or manganese) in conserved site inside barrel
N-terminal alpha+beta domain is common to this superfamily
Family c.1.11.2: D-glucarate dehydratase-like [51609] (15 proteins) 
Protein automated matches [226997] (13 species)
not a true protein
Species Agrobacterium tumefaciens [TaxId:176299] [226506] (6 PDB entries) 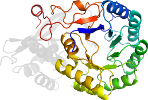
Domain d4hcla2: 4hcl A:127-382 [222503]
Other proteins in same PDB: d4hcla1, d4hcla3, d4hclb1, d4hclb3
automated match to d1rvka1
complexed with llh, mg, pg4
Details for d4hcla2
PDB Entry: 4hcl (more details), 1.8 Å
PDB Description: crystal structure of d-glucarate dehydratase from agrobacterium tumefaciens complexed with magnesium and l-lyxarohydroxamate
PDB Compounds: (A:) isomerase/lactonizing enzymeSCOPe Domain Sequences for d4hcla2:
Sequence; same for both SEQRES and ATOM records: (download)
>d4hcla2 c.1.11.2 (A:127-382) automated matches {Agrobacterium tumefaciens [TaxId: 176299]}
yrdkvlaygsimcgdelegglatpedygrfaetlvkrgykgiklhtwmppvswapdvkmd
lkacaavreavgpdirlmidafhwysrtdalalgrgleklgfdwieepmdeqslssykwl
sdnldipvvgpesaagkhwhraewikagacdilrtgvndvggitpalktmhlaeafgmec
evhgntamnlhvvaatkncrwyergllhpfleyddghdylkslsdpmdrdgfvhvpdrpg
lgedidftfidnnrvr
SCOPe Domain Coordinates for d4hcla2:
Click to download the PDB-style file with coordinates for d4hcla2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d4hcla2:
- d4hcla2 first appeared in SCOPe 2.03
- d4hcla2 appears in SCOPe 2.07

 View in 3D
View in 3D