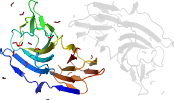Lineage for d3esgb_ (3esg B:)
- Root: SCOPe 2.01

Class b: All beta proteins [48724] (174 folds) 
Fold b.82: Double-stranded beta-helix [51181] (7 superfamilies)
one turn of helix is made by two pairs of antiparallel strands linked with short turns
has appearance of a sandwich of distinct architecture and jelly-roll topology
Superfamily b.82.1: RmlC-like cupins [51182] (25 families) 

Family b.82.1.0: automated matches [191354] (1 protein)
not a true family
Protein automated matches [190388] (5 species)
not a true protein
Species Pseudomonas fluorescens [TaxId:216595] [189073] (1 PDB entry) 
Domain d3esgb_: 3esg B: [175192]
automated match to d1ylla1
complexed with fmt, gol
Details for d3esgb_
PDB Entry: 3esg (more details), 1.8 Å
PDB Description: crystal structure of hutd from pseudomonas fluorescens sbw25
PDB Compounds: (B:) Putative uncharacterized proteinSCOPe Domain Sequences for d3esgb_:
Sequence; same for both SEQRES and ATOM records: (download)
>d3esgb_ b.82.1.0 (B:) automated matches {Pseudomonas fluorescens [TaxId: 216595]}
saisvwravdyvrmpwkngggsteeitrdagtglegfgwrlsiadigesggfssfagyqr
vitviqgagmvltvdgeeqrgllplqpfafrgdsqvscrlitgpirdfnliysperyhar
lqwvdgvqrffstaqtvlvfsvadevkvlgeklghhdclqvdgnaglldisvtgrcclie
ltqrg
SCOPe Domain Coordinates for d3esgb_:
Click to download the PDB-style file with coordinates for d3esgb_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d3esgb_:
- d3esgb_ is new in SCOPe 2.01-stable
- d3esgb_ appears in SCOPe 2.02
- d3esgb_ last appears in SCOPe 2.06

 View in 3D
View in 3D