Lineage for d2gtia2 (2gti A:217-369)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.294: EndoU-like [142876] (1 superfamily)
comprises several helices and two three-stranded antiparallel beta-sheets; similar architecture to the RNase A-like fold (54075)
Superfamily d.294.1: EndoU-like [142877] (3 families) 
similarity to the RNase A-like superfamily (54076) extends to the active site location and architecture; the two structural cores of the RNase A and EndoU superfamilies are interrelated by a topological permutation - transposition of two pereferial beta-strands, suggesting possible distant homology of the two superfamilies (and their unification in a hyperfamily)
Family d.294.1.2: Nsp15 C-terminal domain-like [142881] (2 proteins)
PfamB PB001946
Protein Nsp15, C-terminal domain [142882] (2 species) 
Species Murine hepatitis virus, MHV [TaxId:11138] [142884] (2 PDB entries)
Uniprot P16342 6720-6872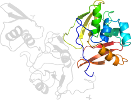
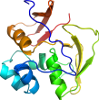
Domain d2gtia2: 2gti A:217-369 [135653]
Other proteins in same PDB: d2gtia1, d2gtia3
complexed with gol, so4; mutant
Details for d2gtia2
PDB Entry: 2gti (more details), 2.15 Å
PDB Description: mutation of mhv coronavirus non-structural protein nsp15 (f307l)
PDB Compounds: (A:) Replicase polyprotein 1abSCOPe Domain Sequences for d2gtia2:
Sequence; same for both SEQRES and ATOM records: (download)
>d2gtia2 d.294.1.2 (A:217-369) Nsp15, C-terminal domain {Murine hepatitis virus, MHV [TaxId: 11138]}
argtiftqsrllssftprsemekdfmdldddvfiakyslqdyafehvvygsfnqkiiggl
hlliglarrqqksnlviqefvtydssihsylitdensgssksvctvidlllddfvdivks
lnlkcvskvvnvnvdfkdfqfmlwcneekvmtf
SCOPe Domain Coordinates for d2gtia2:
Click to download the PDB-style file with coordinates for d2gtia2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d2gtia2:
- d2gtia2 first appeared in SCOP 1.73
- d2gtia2 appears in SCOPe 2.07
 View in 3D
View in 3D
