Lineage for d1zrda2 (1zrd A:8-137)
- Root: SCOP 1.73

Class b: All beta proteins [48724] (165 folds) 
Fold b.82: Double-stranded beta-helix [51181] (7 superfamilies)
one turn of helix is made by two pairs of antiparallel strands linked with short turns
has appearance of a sandwich of distinct architecture and jelly-roll topology
Superfamily b.82.3: cAMP-binding domain-like [51206] (3 families) 

Family b.82.3.2: cAMP-binding domain [51210] (10 proteins)
Pfam PF00027
Protein Catabolite gene activator protein, N-terminal domain [51211] (1 species) 
Species Escherichia coli [TaxId:562] [51212] (19 PDB entries) 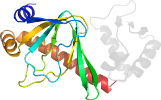
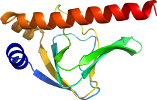
Domain d1zrda2: 1zrd A:8-137 [125536]
Other proteins in same PDB: d1zrda1, d1zrdb1
automatically matched to d1i5zb2
complexed with cmp
Details for d1zrda2
PDB Entry: 1zrd (more details), 2.8 Å
PDB Description: 4 crystal structures of cap-dna with all base-pair substitutions at position 6, cap-[6a;17t]icap38 dna
PDB Compounds: (A:) Catabolite gene activatorSCOP Domain Sequences for d1zrda2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1zrda2 b.82.3.2 (A:8-137) Catabolite gene activator protein, N-terminal domain {Escherichia coli [TaxId: 562]}
dptlewflshchihkypskstlihqgekaetlyyivkgsvavlikdeegkemilsylnqg
dfigelglfeegqersawvraktacevaeisykkfrqliqvnpdilmrlsaqmarrlqvt
sekvgnlafl
SCOP Domain Coordinates for d1zrda2:
Click to download the PDB-style file with coordinates for d1zrda2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1zrda2:
- d1zrda2 is new in SCOP 1.73
- d1zrda2 appears in SCOP 1.75
- d1zrda2 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D