Lineage for d1sxir_ (1sxi R:)
- Root: SCOPe 2.06

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.93: Periplasmic binding protein-like I [53821] (1 superfamily)
consists of two similar intertwined domain with 3 layers (a/b/a) each: duplication
parallel beta-sheet of 6 strands, order 213456
Superfamily c.93.1: Periplasmic binding protein-like I [53822] (2 families) 
Similar in architecture to the superfamily II but partly differs in topology
Family c.93.1.1: L-arabinose binding protein-like [53823] (18 proteins) 
Protein Glucose-resistance amylase regulator CcpA, C-terminal domain [117740] (2 species) 
Species Bacillus megaterium [TaxId:1404] [117741] (10 PDB entries)
Uniprot P46828 58-322 ! Uniprot P46828
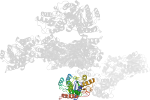
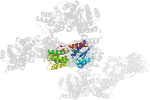
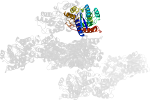
Domain d1sxir_: 1sxi R: [112158]
complexed with mg
Details for d1sxir_
PDB Entry: 1sxi (more details), 3 Å
PDB Description: Structure of apo transcription regulator B. megaterium
PDB Compounds: (R:) Glucose-resistance amylase regulatorSCOPe Domain Sequences for d1sxir_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1sxir_ c.93.1.1 (R:) Glucose-resistance amylase regulator CcpA, C-terminal domain {Bacillus megaterium [TaxId: 1404]}
tttvgviipdisnifyaelargiediatmykyniilsnsdqnqdkelhllnnmlgkqvdg
iifmsgnvteehveelkkspvpvvlaasiestnqipsvtidyeqaafdavqslidsghkn
iafvsgtleepinhakkvkgykraltesglpvrdsyivegdytydsgieaveklleedek
ptaifvgtdemalgvihgaqdrglnvpndleiigfdntrlstmvrpqltsvvqpmydiga
vamrlltkymnketvdssivqlphriefrqstk
SCOPe Domain Coordinates for d1sxir_:
Click to download the PDB-style file with coordinates for d1sxir_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1sxir_:
- d1sxir_ first appeared in SCOP 1.71
- d1sxir_ appears in SCOPe 2.05
- d1sxir_ appears in SCOPe 2.07
- d1sxir_ appears in the current release, SCOPe 2.08

 View in 3D
View in 3D