Lineage for d1vema1 (1vem A:418-516)
- Root: SCOPe 2.04

Class b: All beta proteins [48724] (176 folds) 
Fold b.3: Prealbumin-like [49451] (7 superfamilies)
sandwich; 7 strands in 2 sheets, greek-key
variations: some members have additional 1-2 strands to common fold
Superfamily b.3.1: Starch-binding domain-like [49452] (4 families) 

Family b.3.1.1: Starch-binding domain [49453] (3 proteins)
automatically mapped to Pfam PF00686
Protein beta-amylase [49462] (1 species) 
Species Bacillus cereus [TaxId:1396] [49463] (12 PDB entries) 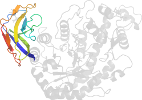
Domain d1vema1: 1vem A:418-516 [120021]
Other proteins in same PDB: d1vema2
automated match to d1j18a1
complexed with ca
Details for d1vema1
PDB Entry: 1vem (more details), 1.85 Å
PDB Description: crystal structure analysis of bacillus cereus beta-amylase at the optimum ph (6.5)
PDB Compounds: (A:) beta-amylaseSCOPe Domain Sequences for d1vema1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1vema1 b.3.1.1 (A:418-516) beta-amylase {Bacillus cereus [TaxId: 1396]}
tpvmqtivvknvpttigdtvyitgnraelgswdtkqypiqlyydshsndwrgnvvlpaer
niefkafikskdgtvkswqtiqqswnpvplkttshtssw
SCOPe Domain Coordinates for d1vema1:
Click to download the PDB-style file with coordinates for d1vema1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1vema1:
- d1vema1 first appeared in SCOP 1.73
- d1vema1 appears in SCOPe 2.03
- d1vema1 appears in SCOPe 2.05
- d1vema1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D