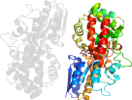
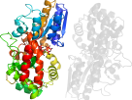Lineage for d1urda_ (1urd A:)
- Root: SCOP 1.71

Class c: Alpha and beta proteins (a/b) [51349] (134 folds) 
Fold c.94: Periplasmic binding protein-like II [53849] (1 superfamily)
consists of two similar intertwined domain with 3 layers (a/b/a) each: duplication
mixed beta-sheet of 5 strands, order 21354; strand 5 is antiparallel to the rest
Superfamily c.94.1: Periplasmic binding protein-like II [53850] (2 families) 
Similar in architecture to the superfamily I but partly differs in topology
Family c.94.1.1: Phosphate binding protein-like [53851] (29 proteins) 
Protein D-maltodextrin-binding protein, MBP [53862] (4 species)
contains a few additional helices in the C-terminal extension; homologous to thiaminase I
Species Alicyclobacillus acidocaldarius [TaxId:405212] [102690] (3 PDB entries) 
Domain d1urda_: 1urd A: [99825]
Details for d1urda_
PDB Entry: 1urd (more details), 1.53 Å
PDB Description: x-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium alicyclobacillus acidocaldarius provide insight into acid stability of proteins
SCOP Domain Sequences for d1urda_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1urda_ c.94.1.1 (A:) D-maltodextrin-binding protein, MBP {Alicyclobacillus acidocaldarius}
qtitvwswqtgpelqdvkqiaaqwakahgdkvivvdqssnpkgfqfyataartgkgpdvv
fgmphdnngvfaeeglmapvpsgvlntglyapntidaikvngtmysvpvsvqvaaiyynk
klvpqppqtwaefvkdanahgfmydqanlyfdyaiiggyggyvfkdnngtldpnnigldt
pgavqaytlmrdmvskyhwmtpstngsiakaeflagkigmyvsgpwdtadiekakidfgv
tpwptlpngkhatpflgvitafvnkesktqaadwslvqaltsaqaqqmyfrdsqqipall
svqrssavqssptfkafveqlryavpmpnipqmqavwqamsilqniiagkvspeqgakdf
vqniqkgima
SCOP Domain Coordinates for d1urda_:
Click to download the PDB-style file with coordinates for d1urda_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1urda_:
- d1urda_ first appeared in SCOP 1.67
- d1urda_ appears in SCOP 1.69
- d1urda_ appears in SCOP 1.73
- d1urda_ appears in the current release, SCOPe 2.08

 View in 3D
View in 3D