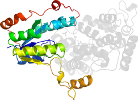
Lineage for d1owla1 (1owl A:205-475)
- Root: SCOPe 2.06

Class a: All alpha proteins [46456] (289 folds) 
Fold a.99: Cryptochrome/photolyase FAD-binding domain [48172] (1 superfamily)
multihelical; consists of two all-alpha subdomains
Superfamily a.99.1: Cryptochrome/photolyase FAD-binding domain [48173] (2 families) 
automatically mapped to Pfam PF03441
Family a.99.1.1: Cryptochrome/photolyase FAD-binding domain [48174] (3 proteins) 
Protein C-terminal domain of DNA photolyase [48175] (3 species)
N-terminal domain is alpha/beta and binds a light-harvesting cofactor
Species Synechococcus elongatus [TaxId:32046] [48177] (7 PDB entries)
Uniprot P05327 1-474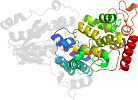
Domain d1owla1: 1owl A:205-475 [93647]
Other proteins in same PDB: d1owla2
complexed with fad, po4
Details for d1owla1
PDB Entry: 1owl (more details), 1.8 Å
PDB Description: Structure of apophotolyase from Anacystis nidulans
PDB Compounds: (A:) Deoxyribodipyrimidine photolyaseSCOPe Domain Sequences for d1owla1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1owla1 a.99.1.1 (A:205-475) C-terminal domain of DNA photolyase {Synechococcus elongatus [TaxId: 32046]}
pvepgetaaiarlqefcdraiadydpqrnfpaeagtsglspalkfgaigirqawqaasaa
halsrsdearnsirvwqqelawrefyqhalyhfpsladgpyrslwqqfpwenrealftaw
tqaqtgypivdaamrqltetgwmhnrcrmivasfltkdliidwrrgeqffmqhlvdgdla
annggwqwsassgmdpkplrifnpasqakkfdatatyikrwlpelrhvhpkdlisgeitp
ierrgypapivnhnlrqkqfkalynqlkaai
SCOPe Domain Coordinates for d1owla1:
Click to download the PDB-style file with coordinates for d1owla1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1owla1:
- d1owla1 first appeared in SCOP 1.67
- d1owla1 appears in SCOPe 2.05
- d1owla1 appears in SCOPe 2.07
- d1owla1 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D