Lineage for d1n9gf1 (1n9g F:23-160,F:350-386)
- Root: SCOP 1.71

Class b: All beta proteins [48724] (149 folds) 
Fold b.35: GroES-like [50128] (2 superfamilies)
contains barrel, partly opened; n*=4, S*=8; meander
Superfamily b.35.1: GroES-like [50129] (2 families) 

Family b.35.1.2: Alcohol dehydrogenase-like, N-terminal domain [50136] (15 proteins)
C-terminal domain is alpha/beta (classical Rossmann-fold)
Protein 2,4-dienoyl-CoA reductase [89309] (1 species) 
Species Yeast (Candida tropicalis) [TaxId:5482] [89310] (5 PDB entries) 
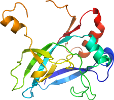
Domain d1n9gf1: 1n9g F:23-160,F:350-386 [91730]
Other proteins in same PDB: d1n9ga2, d1n9gb2, d1n9gc2, d1n9gd2, d1n9ge2, d1n9gf2
complexed with nap, so4
Details for d1n9gf1
PDB Entry: 1n9g (more details), 1.98 Å
PDB Description: mitochondrial 2-enoyl thioester reductase etr1p/etr2p heterodimer from candida tropicalis
SCOP Domain Sequences for d1n9gf1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1n9gf1 b.35.1.2 (F:23-160,F:350-386) 2,4-dienoyl-CoA reductase {Yeast (Candida tropicalis)}
mitaqavlytqhgepkdvlftqsfeidddnlapnevivktlgspinpsdinqiqgvypsk
pakttgfgtaepaapcgneglfevikvgsnvssleagdwvipshvnfgtwrthalgnddd
fiklpnpaqskangkpngXltdaksietlydgtkplhelyqdgvanskdgkqlity
SCOP Domain Coordinates for d1n9gf1:
Click to download the PDB-style file with coordinates for d1n9gf1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1n9gf1:
- d1n9gf1 first appeared in SCOP 1.67
- d1n9gf1 appears in SCOP 1.69
- d1n9gf1 appears in SCOP 1.73
- d1n9gf1 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D