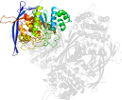
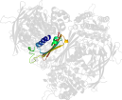
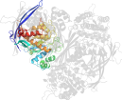
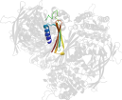
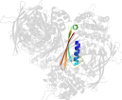
Lineage for d1p1hc2 (1p1h C:323-437)
- Root: SCOP 1.69

Class d: Alpha and beta proteins (a+b) [53931] (279 folds) 
Fold d.81: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain [55346] (1 superfamily)
core: alpha-beta-alpha-beta(3); mixed sheet: 2134, strand 2 is parallel to strand 1
Superfamily d.81.1: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain [55347] (5 families) 
N-terminal domain is the classic Rossmann-fold
Family d.81.1.3: Dihydrodipicolinate reductase-like [55368] (6 proteins) 
Protein Myo-inositol 1-phosphate synthase [75484] (4 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [75486] (9 PDB entries) 
Domain d1p1hc2: 1p1h C:323-437 [87680]
Other proteins in same PDB: d1p1ha1, d1p1hb1, d1p1hc1, d1p1hd1
Details for d1p1hc2
PDB Entry: 1p1h (more details), 1.95 Å
PDB Description: Crystal structure of the 1L-myo-inositol/NAD+ complex
SCOP Domain Sequences for d1p1hc2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1p1hc2 d.81.1.3 (C:323-437) Myo-inositol 1-phosphate synthase {Baker's yeast (Saccharomyces cerevisiae)}
sgqtklksvlaqflvdagikpvsiasynhlgnndgynlsapkqfrskeiskssviddiia
sndilyndklgkkvdhcivikymkpvgdskvamdeyyselmlgghnrisihnvce
SCOP Domain Coordinates for d1p1hc2:
Click to download the PDB-style file with coordinates for d1p1hc2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1p1hc2:
- d1p1hc2 first appeared in SCOP 1.65
- d1p1hc2 appears in SCOP 1.67
- d1p1hc2 appears in SCOP 1.71
- d1p1hc2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D