Lineage for d1p16a2 (1p16 A:1-245)
- Root: SCOPe 2.04

Class d: Alpha and beta proteins (a+b) [53931] (380 folds) 
Fold d.142: ATP-grasp [56058] (2 superfamilies)
Consists of two subdomains with different alpha+beta folds
shares functional and structural similarities with the PIPK and protein kinase superfamilies
Superfamily d.142.2: DNA ligase/mRNA capping enzyme, catalytic domain [56091] (6 families) 
has a circularly permuted topology
Family d.142.2.3: mRNA capping enzyme [56100] (2 proteins)
automatically mapped to Pfam PF01331
Protein mRNA capping enzyme alpha subunit [90032] (1 species) 
Species Yeast (Candida albicans) [TaxId:5476] [90033] (1 PDB entry) 
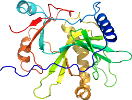
Domain d1p16a2: 1p16 A:1-245 [87662]
Other proteins in same PDB: d1p16a1, d1p16b1
complexed with the phosphorylated carboxyl-terminal peptide of RNA polymerase II, chains C and D
complexed with g, gtp, po4
Details for d1p16a2
PDB Entry: 1p16 (more details), 2.7 Å
PDB Description: structure of an mrna capping enzyme bound to the phosphorylated carboxyl-terminal domain of rna polymerase ii
PDB Compounds: (A:) mRNA capping enzyme alpha subunitSCOPe Domain Sequences for d1p16a2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1p16a2 d.142.2.3 (A:1-245) mRNA capping enzyme alpha subunit {Yeast (Candida albicans) [TaxId: 5476]}
mvqleereipvipgnkldeeetkelrlmvaellgrrntgfpgsqpvsferrhleetlmqk
dyfvcektdglrcllflindpdkgegvflvtrendyyfipnihfplsvnetrekptyhhg
tlldgelvlenrnvsepvlryvifdalaihgkciidrplpkrlgyitenvmkpfdnfkkh
npdivnspefpfkvgfktmltsyhaddvlskmdklfhasdgliytcaetpyvfgtdqtll
kwkpa
SCOPe Domain Coordinates for d1p16a2:
Click to download the PDB-style file with coordinates for d1p16a2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1p16a2:
- d1p16a2 first appeared in SCOP 1.65
- d1p16a2 appears in SCOPe 2.03
- d1p16a2 appears in SCOPe 2.05
- d1p16a2 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D


