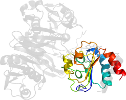
Lineage for d1o97d2 (1o97 D:196-318)
- Root: SCOPe 2.05

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.31: DHS-like NAD/FAD-binding domain [52466] (1 superfamily)
3 layers: a/b/a; parallel beta-sheet of 6 strands, order 321456; Rossmann-like
Superfamily c.31.1: DHS-like NAD/FAD-binding domain [52467] (7 families) 
binds cofactor molecules in the opposite direction than classical Rossmann fold
Family c.31.1.2: C-terminal domain of the electron transfer flavoprotein alpha subunit [52471] (1 protein)
lacks strand 3; shares the FAD-binding mode with the pyruvate oxidase domain
Protein C-terminal domain of the electron transfer flavoprotein alpha subunit [52472] (3 species) 
Species Methylophilus methylotrophus [TaxId:17] [82379] (6 PDB entries) 
Domain d1o97d2: 1o97 D:196-318 [81234]
Other proteins in same PDB: d1o97c_, d1o97d1
complexed with amp, fad
Details for d1o97d2
PDB Entry: 1o97 (more details), 1.6 Å
PDB Description: structure of electron transferring flavoprotein from methylophilus methylotrophus, recognition loop removed by limited proteolysis
PDB Compounds: (D:) electron transferring flavoprotein alpha-subunitSCOPe Domain Sequences for d1o97d2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1o97d2 c.31.1.2 (D:196-318) C-terminal domain of the electron transfer flavoprotein alpha subunit {Methylophilus methylotrophus [TaxId: 17]}
didittvdfimsigrgigeetnveqfreladeagatlccsrpiadagwlpksrqvgqsgk
vvgscklyvamgisgsiqhmagmkhvptiiavntdpgasiftiakygivadifdieeelk
aql
SCOPe Domain Coordinates for d1o97d2:
Click to download the PDB-style file with coordinates for d1o97d2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1o97d2:
- d1o97d2 first appeared in SCOP 1.63
- d1o97d2 appears in SCOPe 2.04
- d1o97d2 appears in SCOPe 2.06
- d1o97d2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D