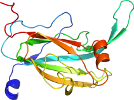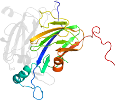Lineage for d1l9sc1 (1l9s C:4-166)
- Root: SCOP 1.69

Class b: All beta proteins [48724] (144 folds) 
Fold b.6: Cupredoxin-like [49502] (2 superfamilies)
sandwich; 7 strands in 2 sheets, greek-key
variations: some members have additional 1-2 strands
Superfamily b.6.1: Cupredoxins [49503] (6 families) 
contains copper-binding site
Family b.6.1.3: Multidomain cupredoxins [49550] (7 proteins) 
Protein Nitrite reductase, NIR [49551] (5 species)
consists of two domains of this fold
Species Alcaligenes faecalis, strain s-6 [TaxId:511] [49553] (23 PDB entries) 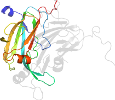
Domain d1l9sc1: 1l9s C:4-166 [77856]
Details for d1l9sc1
PDB Entry: 1l9s (more details), 1.78 Å
PDB Description: crystal structure of the i257t variant of the copper-containing nitrite reductase from alcaligenes faecalis s-6
SCOP Domain Sequences for d1l9sc1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1l9sc1 b.6.1.3 (C:4-166) Nitrite reductase, NIR {Alcaligenes faecalis, strain s-6}
ataaeiaalprqkvelvdppfvhahsqvaeggpkvveftmvieekkividdagtevhama
fngtvpgplmvvhqddyleltlinpetntlmhnidfhaatgalgggglteinpgektilr
fkatkpgvfvyhcappgmvpwhvvsgmngaimvlpreglhdgk
SCOP Domain Coordinates for d1l9sc1:
Click to download the PDB-style file with coordinates for d1l9sc1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1l9sc1:
- d1l9sc1 first appeared in SCOP 1.63
- d1l9sc1 appears in SCOP 1.67
- d1l9sc1 appears in SCOP 1.71
- d1l9sc1 appears in the current release, SCOPe 2.08
 View in 3D View in 3DDomains from other chains: (mouse over for more information) d1l9sa1, d1l9sa2, d1l9sb1, d1l9sb2 |