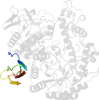Lineage for d1kwga3 (1kwg A:394-590)
- Root: SCOPe 2.05

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.23: Flavodoxin-like [52171] (15 superfamilies)
3 layers, a/b/a; parallel beta-sheet of 5 strand, order 21345
Superfamily c.23.16: Class I glutamine amidotransferase-like [52317] (10 families) 
conserved positions of the oxyanion hole and catalytic nucleophile; different constituent families contain different additional structures
Family c.23.16.5: A4 beta-galactosidase middle domain [82351] (1 protein)
probable non-catalytic branch of the class I GAT family; overall fold is very similar but the active site is not conserved
automatically mapped to Pfam PF08532
Protein A4 beta-galactosidase middle domain [82352] (1 species) 
Species Thermus thermophilus [TaxId:274] [82353] (2 PDB entries) 
Domain d1kwga3: 1kwg A:394-590 [77563]
Other proteins in same PDB: d1kwga1, d1kwga2
complexed with act, cl, mpd, zn
Details for d1kwga3
PDB Entry: 1kwg (more details), 1.6 Å
PDB Description: Crystal structure of Thermus thermophilus A4 beta-galactosidase
PDB Compounds: (A:) beta-galactosidaseSCOPe Domain Sequences for d1kwga3:
Sequence; same for both SEQRES and ATOM records: (download)
>d1kwga3 c.23.16.5 (A:394-590) A4 beta-galactosidase middle domain {Thermus thermophilus [TaxId: 274]}
pvaqapvalvfdyeaawiyevqpqgaewsylglvylfysalrrlgldvdvvppgaslrgy
afavvpslpivreealeafreaegpvlfgprsgsktetfqipkelppgplqallplkvvr
veslppgllevaegalgrfplglwrewveaplkplltfqdgkgalyregrylylaawpsp
elagrllsalaaeaglk
SCOPe Domain Coordinates for d1kwga3:
Click to download the PDB-style file with coordinates for d1kwga3.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1kwga3:
- d1kwga3 first appeared in SCOP 1.63
- d1kwga3 appears in SCOPe 2.04
- d1kwga3 appears in SCOPe 2.06
- d1kwga3 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D