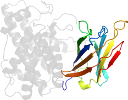Lineage for d1gyga1 (1gyg A:1-249)
- Root: SCOPe 2.06

Class a: All alpha proteins [46456] (289 folds) 
Fold a.124: Phospholipase C/P1 nuclease [48536] (1 superfamily)
multihelical
Superfamily a.124.1: Phospholipase C/P1 nuclease [48537] (3 families) 
duplication: all chain but the N-terminal helix forms two structural repeats
Family a.124.1.1: Phospholipase C [48538] (3 proteins) 
Protein Alpha-toxin, N-terminal domain [48541] (2 species) 
Species Clostridium perfringens, different strains [TaxId:1502] [48542] (6 PDB entries) 
Domain d1gyga1: 1gyg A:1-249 [70753]
Other proteins in same PDB: d1gyga2, d1gygb2
complexed with zn
Details for d1gyga1
PDB Entry: 1gyg (more details), 1.9 Å
PDB Description: r32 closed form of alpha-toxin from clostridium perfringens strain cer89l43
PDB Compounds: (A:) phospholipase cSCOPe Domain Sequences for d1gyga1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1gyga1 a.124.1.1 (A:1-249) Alpha-toxin, N-terminal domain {Clostridium perfringens, different strains [TaxId: 1502]}
wdgkidgtgthamivtqgvsilendlsknepesvrknleilkenmhelqlgstypdydkn
aydlyqdhfwdpdtdnnfskdnswylaysipdtgesqirkfsalaryewqrgnykqatfy
lgeamhyfgdidtpyhpanvtavdsaghvkfetfaeerkeqykintvgcktnedfyadil
knkdfnawskeyargfaktgksiyyshasmshswddwdyaakvtlansqkgtagyiyrfl
hdvsegndp
SCOPe Domain Coordinates for d1gyga1:
Click to download the PDB-style file with coordinates for d1gyga1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1gyga1:
- d1gyga1 first appeared in SCOP 1.61
- d1gyga1 appears in SCOPe 2.05
- d1gyga1 appears in SCOPe 2.07
- d1gyga1 appears in the current release, SCOPe 2.08
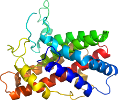
 View in 3D
View in 3D