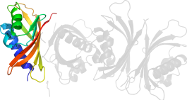
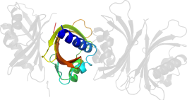
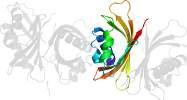Lineage for d1gyba_ (1gyb A:)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.17: Cystatin-like [54402] (7 superfamilies)
Core: alpha-beta(4); helix packs against coiled antiparallel beta-sheet
Superfamily d.17.4: NTF2-like [54427] (31 families) 
has a beta-alpha(2)-beta insertion after the main helix
Family d.17.4.2: NTF2-like [54431] (6 proteins) 
Protein Nuclear transport factor-2 (NTF2) [54432] (4 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [75374] (2 PDB entries) 
Domain d1gyba_: 1gyb A: [70744]
complexed with FxFG nucleoporin repeat
mutant
Details for d1gyba_
PDB Entry: 1gyb (more details), 1.9 Å
PDB Description: n77y point mutant of yntf2 bound to fxfg nucleoporin repeat
PDB Compounds: (A:) nuclear transport factor 2SCOPe Domain Sequences for d1gyba_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1gyba_ d.17.4.2 (A:) Nuclear transport factor-2 (NTF2) {Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]}
ldfntlaqnftqfyynqfdtdrsqlgnlyrnesmltfetsqlqgakdiveklvslpfqkv
qhrittldaqpaspygdvlvmitgdllideeqnpqrfsqvfhlipdgnsyyvfndifrln
ys
SCOPe Domain Coordinates for d1gyba_:
Click to download the PDB-style file with coordinates for d1gyba_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1gyba_:
- d1gyba_ first appeared in SCOP 1.61
- d1gyba_ appears in SCOPe 2.07

 View in 3D
View in 3D