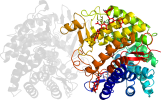Lineage for d1krfa_ (1krf A:)
- Root: SCOPe 2.07

Class a: All alpha proteins [46456] (289 folds) 
Fold a.102: alpha/alpha toroid [48207] (6 superfamilies)
multihelical; up to seven alpha-hairpins are arranged in closed circular array; there may be sequence similarities between different superfamilies
Superfamily a.102.2: Seven-hairpin glycosidases [48225] (1 family) 
automatically mapped to Pfam PF01532
Family a.102.2.1: Class I alpha-1;2-mannosidase, catalytic domain [48226] (1 protein) 
Protein Class I alpha-1;2-mannosidase, catalytic domain [48227] (5 species) 
Species Fungus (Penicillium citrinum) [TaxId:5077] [69088] (5 PDB entries) 
Domain d1krfa_: 1krf A: [68850]
complexed with ca, kif
Details for d1krfa_
PDB Entry: 1krf (more details), 2.2 Å
PDB Description: structure of p. citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the er and golgi class i enzymes
PDB Compounds: (A:) Mannosyl-oligosaccharide alpha-1,2-mannosidaseSCOPe Domain Sequences for d1krfa_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1krfa_ a.102.2.1 (A:) Class I alpha-1;2-mannosidase, catalytic domain {Fungus (Penicillium citrinum) [TaxId: 5077]}
snqakadavkeafqhawngymkyafphdeltpvsnghadsrngwgasavdalstavimgk
advvnailehvadidfsktsdtvslfettirylagmlsgydllqgpaknlvdnqdlidgl
ldqsrnladvlkfafdtpsgvpynninitshgndgattnglavtgtlvlewtrlsdltgd
eeyaklsqkaesyllkpqpsssepfpglvgssinindgqfadsrvswnggddsfyeylik
myvydpkrfetykdrwvlaaestikhlkshpksrpdltflssysnrnydlssqhltcfdg
gsfllggtvldrqdfidfglelvdgceatynstltkigpdswgwdpkkvpsdqkefyeka
gfyissgsyvlrpeviesfyyahrvtgkeiyrdwvwnafvainstcrtdsgfaavsdvnk
anggskydnqesflfaevmkysylahsedaawqvqkggkntfvynteahpisvar
SCOPe Domain Coordinates for d1krfa_:
Click to download the PDB-style file with coordinates for d1krfa_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1krfa_:
- d1krfa_ first appeared in SCOP 1.59
- d1krfa_ appears in SCOPe 2.06
- d1krfa_ appears in the current release, SCOPe 2.08
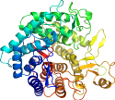
 View in 3D
View in 3D