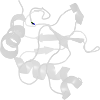Lineage for d1jrma1 (1jrm A:2-104)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.206: YggU-like [69785] (1 superfamily)
beta(2)-loop-alpha-beta(2)-alpha; 2 layers: a/b; antiparallel sheet 1243; some similarity to the Homing endonuclease-like fold
Superfamily d.206.1: YggU-like [69786] (1 family) 

Family d.206.1.1: YggU-like [69787] (2 proteins) 
Protein Hypothetical protein MTH637 [69788] (1 species) 
Species Methanobacterium thermoautotrophicum [TaxId:145262] [69789] (1 PDB entry) 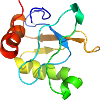
Domain d1jrma1: 1jrm A:2-104 [67136]
Other proteins in same PDB: d1jrma2
structural genomics
Details for d1jrma1
PDB Entry: 1jrm (more details)
PDB Description: nmr structure of mth0637. ontario centre for structural proteomics target mth0637_1_104; northeast structural genomics target tt135
PDB Compounds: (A:) CONSERVED HYPOTHETICAL PROTEIN mth637SCOPe Domain Sequences for d1jrma1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1jrma1 d.206.1.1 (A:2-104) Hypothetical protein MTH637 {Methanobacterium thermoautotrophicum [TaxId: 145262]}
itmdclrevgddllvnievspasgkfgipsynewrkrievkihsppqkgkanreiikefs
etfgrdveivsgqksrqktiriqgmgrdlflklvsekfgleip
SCOPe Domain Coordinates for d1jrma1:
Click to download the PDB-style file with coordinates for d1jrma1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1jrma1:
- d1jrma1 first appeared in SCOP 1.59, called d1jrma_
- d1jrma1 appears in SCOPe 2.07
 View in 3D
View in 3D