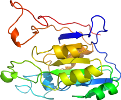
Lineage for d1ieva2 (1iev A:389-602)
- Root: SCOPe 2.06

Class c: Alpha and beta proteins (a/b) [51349] (148 folds) 
Fold c.23: Flavodoxin-like [52171] (15 superfamilies)
3 layers, a/b/a; parallel beta-sheet of 5 strand, order 21345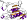
Superfamily c.23.11: Beta-D-glucan exohydrolase, C-terminal domain-like [52279] (2 families) 
automatically mapped to Pfam PF01915
Family c.23.11.1: Beta-D-glucan exohydrolase, C-terminal domain-like [52280] (3 proteins) 
Protein Beta-D-glucan exohydrolase, C-terminal domain [52281] (2 species)
interdomain linker forms an additional, N-terminal strand
Species Barley (Hordeum vulgare) [TaxId:4513] [52282] (9 PDB entries) 
Domain d1ieva2: 1iev A:389-602 [66124]
Other proteins in same PDB: d1ieva1
complexed with ins, nag
Details for d1ieva2
PDB Entry: 1iev (more details), 2.8 Å
PDB Description: crystal structure of barley beta-d-glucan glucohydrolase isoenzyme exo1 in complex with cyclohexitol
PDB Compounds: (A:) beta-d-glucan glucohydrolase isoenzyme exo1SCOPe Domain Sequences for d1ieva2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1ieva2 c.23.11.1 (A:389-602) Beta-D-glucan exohydrolase, C-terminal domain {Barley (Hordeum vulgare) [TaxId: 4513]}
lvllkngktstdapllplpkkapkilvagshadnlgyqcggwtiewqgdtgrttvgttil
eavkaavdpstvvvfaenpdaefvksggfsyaivavgehpytetkgdnlnltipepglst
vqavcggvrcatvlisgrpvvvqpllaasdalvaawlpgsegqgvtdalfgdfgftgrlp
rtwfksvdqlpmnvgdahydplfrlgyglttnat
SCOPe Domain Coordinates for d1ieva2:
Click to download the PDB-style file with coordinates for d1ieva2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1ieva2:
- d1ieva2 first appeared in SCOP 1.59
- d1ieva2 appears in SCOPe 2.05
- d1ieva2 appears in SCOPe 2.07
- d1ieva2 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D