Lineage for d1hr8f2 (1hr8 F:246-462)
- Root: SCOP 1.69

Class d: Alpha and beta proteins (a+b) [53931] (279 folds) 
Fold d.185: LuxS/MPP-like metallohydrolase [63410] (1 superfamily)
core: beta-alpha-beta(2)-alpha(2); 2 layers: alpha/beta
Superfamily d.185.1: LuxS/MPP-like metallohydrolase [63411] (2 families) 
Share the same "active site motif" HxxEH located in the first core helix, but differ in one of the zinc-binding residues
Family d.185.1.1: MPP-like [63412] (4 proteins)
Common fold elaborated with many additional structures; duplication: each family member consists of two similar domains of beta(2)-alpha(2)-beta(2)-alpha(5)-beta structure, but only the N-terminal domain of MPP beta chain binds the catalytic metal
Protein Mitochondrial processing peptidase (MPP) beta chain [64300] (1 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [64301] (4 PDB entries) 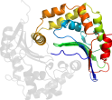
Domain d1hr8f2: 1hr8 F:246-462 [61207]
Other proteins in same PDB: d1hr8a1, d1hr8a2, d1hr8c1, d1hr8c2, d1hr8e1, d1hr8e2, d1hr8g1, d1hr8g2
Details for d1hr8f2
PDB Entry: 1hr8 (more details), 2.7 Å
PDB Description: yeast mitochondrial processing peptidase beta-e73q mutant complexed with cytochrome c oxidase iv signal peptide
SCOP Domain Sequences for d1hr8f2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1hr8f2 d.185.1.1 (F:246-462) Mitochondrial processing peptidase (MPP) beta chain {Baker's yeast (Saccharomyces cerevisiae)}
gplpvfcrgerfikentlptthiaialegvswsapdyfvalatqaivgnwdraigtgtns
psplavaasqngslansymsfstsyadsglwgmyivtdsnehnvrlivneilkewkriks
gkisdaevnrakaqlkaalllsldgstaivedigrqvvttgkrlspeevfeqvdkitkdd
iimwanyrlqnkpvsmvalgntstvpnvsyieeklnq
SCOP Domain Coordinates for d1hr8f2:
Click to download the PDB-style file with coordinates for d1hr8f2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1hr8f2:
- d1hr8f2 first appeared in SCOP 1.57
- d1hr8f2 appears in SCOP 1.67
- d1hr8f2 appears in SCOP 1.71
- d1hr8f2 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D