Lineage for d1ezve2 (1ezv E:31-86)
- Root: SCOP 1.69

Class f: Membrane and cell surface proteins and peptides [56835] (47 folds) 
Fold f.23: Single transmembrane helix [81407] (30 superfamilies)
not a true fold
Superfamily f.23.12: ISP transmembrane anchor [81502] (1 family) 

Family f.23.12.1: ISP transmembrane anchor [81501] (2 proteins) 
Protein Iron-sulfur subunit (ISP) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane region [81500] (3 species) 
Species Baker's yeast (Saccharomyces cerevisiae) [TaxId:4932] [81499] (4 PDB entries) 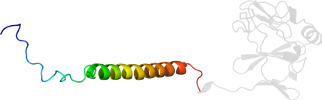
Domain d1ezve2: 1ezv E:31-86 [59549]
Other proteins in same PDB: d1ezva1, d1ezva2, d1ezvb1, d1ezvb2, d1ezvc2, d1ezvc3, d1ezvd1, d1ezvd2, d1ezve1, d1ezvf_, d1ezvg_, d1ezvh_, d1ezvi_, d1ezvx_, d1ezvy_
Details for d1ezve2
PDB Entry: 1ezv (more details), 2.3 Å
PDB Description: structure of the yeast cytochrome bc1 complex co-crystallized with an antibody fv-fragment
SCOP Domain Sequences for d1ezve2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1ezve2 f.23.12.1 (E:31-86) Iron-sulfur subunit (ISP) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane region {Baker's yeast (Saccharomyces cerevisiae)}
kstyrtpnfddvlkenndadkgrsyayfmvgamgllssagakstvetfissmtata
SCOP Domain Coordinates for d1ezve2:
Click to download the PDB-style file with coordinates for d1ezve2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1ezve2:
- d1ezve2 first appeared in SCOP 1.57
- d1ezve2 appears in SCOP 1.67
- d1ezve2 appears in SCOP 1.71
- d1ezve2 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D