Lineage for Species: Escherichia coli [TaxId: 562]
- Root: SCOPe 2.07

Class d: Alpha and beta proteins (a+b) [53931] (388 folds) 
Fold d.68: IF3-like [55199] (8 superfamilies)
beta-alpha-beta-alpha-beta(2); 2 layers; mixed sheet 1243, strand 4 is antiparallel to the rest
Superfamily d.68.2: EPT/RTPC-like [55205] (3 families) 

Family d.68.2.2: Enolpyruvate transferase, EPT [55209] (3 proteins)
duplication: 6 repeats of this fold are organized in two RPTC-like domains
automatically mapped to Pfam PF00275
Protein UDP-N-acetylglucosamine enolpyruvyl transferase (EPT, MurA, MurZ) [55210] (3 species) 
Species Escherichia coli [TaxId:562] [55211] (3 PDB entries)
PDB entries in Species: Escherichia coli [TaxId: 562]:
- Domain(s) for 1a2n:
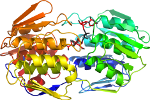
Domain d1a2na_: 1a2n A: [39575]
complexed with tet; mutant
- Domain(s) for 1uae:
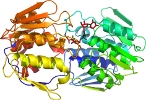
Domain d1uaea_: 1uae A: [39574]
complexed with ffq, ud1
- Domain(s) for 2z2c:

Domain d2z2ca_: 2z2c A: [171009]
automated match to d1dlga_
complexed with udc, udp
Domain d2z2cb_: 2z2c B: [171010]
automated match to d1dlga_
complexed with udc, udp
Domain d2z2cc_: 2z2c C: [171011]
automated match to d1dlga_
complexed with udc, udp
Domain d2z2cd_: 2z2c D: [171012]
automated match to d1dlga_
complexed with udc, udp
More info for Species Escherichia coli [TaxId:562] from d.68.2.2 UDP-N-acetylglucosamine enolpyruvyl transferase (EPT, MurA, MurZ)
Timeline for Species Escherichia coli [TaxId:562] from d.68.2.2 UDP-N-acetylglucosamine enolpyruvyl transferase (EPT, MurA, MurZ):
- Species Escherichia coli [TaxId:562] from d.68.2.2 UDP-N-acetylglucosamine enolpyruvyl transferase (EPT, MurA, MurZ) first appeared (with stable ids) in SCOP 1.55
- Species Escherichia coli [TaxId:562] from d.68.2.2 UDP-N-acetylglucosamine enolpyruvyl transferase (EPT, MurA, MurZ) appears in SCOPe 2.06
- Species Escherichia coli [TaxId:562] from d.68.2.2 UDP-N-acetylglucosamine enolpyruvyl transferase (EPT, MurA, MurZ) appears in the current release, SCOPe 2.08