Lineage for Superfamily b.82.1: RmlC-like cupins
- Root: SCOPe 2.05

Class b: All beta proteins [48724] (176 folds) 
Fold b.82: Double-stranded beta-helix [51181] (7 superfamilies)
one turn of helix is made by two pairs of antiparallel strands linked with short turns
has appearance of a sandwich of distinct architecture and jelly-roll topology
Superfamily b.82.1: RmlC-like cupins [51182] (25 families) 
Families:
b.82.1.1: dTDP-sugar isomerase [51183] (5 proteins) 
b.82.1.2: Germin/Seed storage 7S protein [51187] (5 proteins) 
b.82.1.3: Type I phosphomannose isomerase [51191] (3 proteins)
Share a common two-domain fold with the 7S protein; there is a metal-binding site in the N-terminal domain similar to the metal-binding site of germin
b.82.1.4: Homogentisate dioxygenase [51194] (1 protein)
Share a common two-domain fold with the 7S protein; there is a metal-binding site in the C-terminal domain similar to the metal-binding site of germin
automatically mapped to Pfam PF04209
b.82.1.5: Quercetin 2,3-dioxygenase-like [75035] (3 proteins)
Share a common two-domain fold with the 7S protein; there is a metal-binding site in the N-terminal domain similar to the metal-binding site of germin
b.82.1.6: Acireductone dioxygenase [82191] (2 proteins)
automatically mapped to Pfam PF03079
b.82.1.7: Glucose-6-phosphate isomerase, GPI [89403] (2 proteins)
automatically mapped to Pfam PF06560
b.82.1.8: Hypothetical protein TM1112 [89406] (1 protein) 
b.82.1.9: TM1287-like [89409] (5 proteins) 
b.82.1.10: TM1459-like [101976] (2 proteins) 
b.82.1.11: YlbA-like [101979] (4 proteins)
Pfam PF05899; formerly pfam06038; duplication: consists of two germin-like domains
b.82.1.12: Pirin-like [101984] (3 proteins)
Share a common two-domain fold with the 7S protein; there is a metal-binding site in the N-terminal domain similar to the metal-binding site of germin
b.82.1.13: KduI-like [110318] (1 protein)
Pfam PF04962: duplication: consists of two germin-like domains
b.82.1.14: Ureidoglycolate hydrolase AllA [117312] (1 protein)
Pfam PF04115; beta-hairpin-swapped dimeric protein of the germin-like fold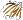
b.82.1.15: Probable transcriptional regulator VC1968, C-terminal domain [117315] (1 protein) 
b.82.1.16: YML079-like [117318] (4 proteins)
Pfam PF06172; DUF985
b.82.1.17: PA5104-like [141609] (2 proteins)
Pfam PF05962; DUF886; duplication: consists of two germin-like domains; overall structural similarity to the YlbA-like family (101979) except a deletion in the interdomain linker region
b.82.1.18: MJ0764-like [141612] (1 protein)
automatically mapped to Pfam PF04962
b.82.1.19: Cysteine dioxygenase type I [141615] (2 proteins)
Pfam PF05995, CDO_I
b.82.1.20: 3-hydroxyanthranilic acid dioxygenase-like [141618] (1 protein)
Pfam PF06052; 3-HAO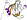
b.82.1.21: Acetylacetone-cleaving enzyme-like [159293] (2 proteins)
PfamB PB026844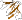
b.82.1.22: VPA0057-like [159296] (1 protein)
Pfam PF06865; DUF1255; non-canonical dimerisation mode
b.82.1.23: Gentisate 1,2-dioxygenase-like [159299] (2 proteins)
Share a common two-domain fold with the 7S protein; there is a metal-binding site in the N-terminal domain similar to the metal-binding site of germin; homotetramer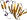
b.82.1.24: EutQ-like [159304] (1 protein)
Pfam PF06249
b.82.1.0: automated matches [191354] (1 protein)
not a true family
More info for Superfamily b.82.1: RmlC-like cupins
Timeline for Superfamily b.82.1: RmlC-like cupins:
- Superfamily b.82.1: RmlC-like cupins first appeared (with stable ids) in SCOP 1.55, called Superfamily b.82.1: RmlC-like
- Superfamily b.82.1: RmlC-like cupins appears in SCOPe 2.04
- Superfamily b.82.1: RmlC-like cupins appears in SCOPe 2.06
- Superfamily b.82.1: RmlC-like cupins appears in the current release, SCOPe 2.08