Lineage for d1rafd2 (1raf D:101-153)
- Root: SCOP 1.71

Class g: Small proteins [56992] (79 folds) 
Fold g.41: Rubredoxin-like [57769] (14 superfamilies)
metal(zinc or iron)-bound fold; sequence contains two CX(n)C motifs, in most cases n = 2
Superfamily g.41.7: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domain [57825] (1 family) 
Family g.41.7.1: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domain [57826] (1 protein) 
Protein Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domain [57827] (2 species) 
Species Escherichia coli [TaxId:562] [57828] (33 PDB entries) 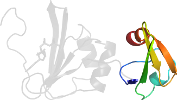
Domain d1rafd2: 1raf D:101-153 [45284]
Other proteins in same PDB: d1rafa1, d1rafa2, d1rafb1, d1rafc1, d1rafc2, d1rafd1
complexed with ctp, zn
Details for d1rafd2
PDB Entry: 1raf (more details), 2.5 Å
PDB Description: crystal structure of ctp-ligated t state aspartate transcarbamoylase at 2.5 angstroms resolution: implications for atcase mutants and the mechanism of negative cooperativity
SCOP Domain Sequences for d1rafd2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1rafd2 g.41.7.1 (D:101-153) Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domain {Escherichia coli}
eridnvlvcpnsncishaepvsssfavrkrandialkckycekefshnvvlan
SCOP Domain Coordinates for d1rafd2:
Click to download the PDB-style file with coordinates for d1rafd2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1rafd2:
- d1rafd2 first appeared (with stable ids) in SCOP 1.55
- d1rafd2 appears in SCOP 1.69
- d1rafd2 appears in SCOP 1.73
- d1rafd2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D