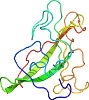
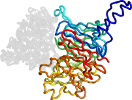
Lineage for d1mael_ (1mae L:)
- Root: SCOP 1.73

Class g: Small proteins [56992] (85 folds) 
Fold g.21: Methylamine dehydrogenase, L chain [57560] (1 superfamily)
disulfide-rich; nearly all-beta
Superfamily g.21.1: Methylamine dehydrogenase, L chain [57561] (1 family) 

Family g.21.1.1: Methylamine dehydrogenase, L chain [57562] (1 protein) 
Protein Methylamine dehydrogenase [57563] (2 species) 
Species Gram negative methylotrophic bacteria (Thiobacillus versutus) [TaxId:34007] [57565] (3 PDB entries) 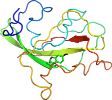
Domain d1mael_: 1mae L: [44889]
Other proteins in same PDB: d1maeh_
complexed with hdz, qtr
Details for d1mael_
PDB Entry: 1mae (more details), 2.8 Å
PDB Description: The Active Site Structure of Methylamine Dehydrogenase: Hydrazines Identify C6 as the Reactive Site of the Tryptophan Derived Quinone Cofactor
PDB Compounds: (L:) methylamine dehydrogenase (light subunit)SCOP Domain Sequences for d1mael_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1mael_ g.21.1.1 (L:) Methylamine dehydrogenase {Gram negative methylotrophic bacteria (Thiobacillus versutus) [TaxId: 34007]}
vdprakwqpqdndiqacdywrhcsidgnicdcsggsltncppgtklataswvascynptd
gqsyliayrdccgynvsgrcpclntegelpvyrpefandiiwcfgaeddamtyhctispi
vgka
SCOP Domain Coordinates for d1mael_:
Click to download the PDB-style file with coordinates for d1mael_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1mael_:
- d1mael_ first appeared (with stable ids) in SCOP 1.55
- d1mael_ appears in SCOP 1.71
- d1mael_ appears in SCOP 1.75
- d1mael_ appears in the current release, SCOPe 2.08
 View in 3D
View in 3D