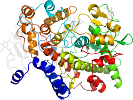
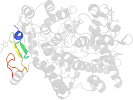Lineage for d1pth_2 (1pth 33-73)
- Root: SCOP 1.55

Class g: Small proteins [56992] (54 folds) 
Fold g.3: Knottins (small inhibitors, toxins, lectins) [57015] (16 superfamilies) 
Superfamily g.3.11: EGF/Laminin [57196] (4 families) 
Family g.3.11.1: EGF-type module [57197] (16 proteins) 
Protein Prostaglandin H2 synthase-1, EGF-like module [57210] (2 species) 
Species Sheep (Ovis aries) [TaxId:9940] [57211] (8 PDB entries) 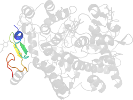
Domain d1pth_2: 1pth 33-73 [44248]
Other proteins in same PDB: d1pth_1
Details for d1pth_2
PDB Entry: 1pth (more details), 3.4 Å
PDB Description: the structural basis of aspirin activity inferred from the crystal structure of inactivated prostaglandin h2 synthase
SCOP Domain Sequences for d1pth_2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1pth_2 g.3.11.1 (33-73) Prostaglandin H2 synthase-1, EGF-like module {Sheep (Ovis aries)}
vnpccyypcqhqgicvrfgldryqcdctrtgysgpnctipe
SCOP Domain Coordinates for d1pth_2:
Click to download the PDB-style file with coordinates for d1pth_2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1pth_2:
- d1pth_2 appears in SCOP 1.57
- d1pth_2 appears in the current release, SCOPe 2.08, called d1ptha2

 View in 3D
View in 3D