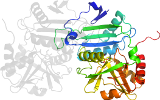Lineage for d1a0ga_ (1a0g A:)
- Root: SCOPe 2.06

Class e: Multi-domain proteins (alpha and beta) [56572] (69 folds) 
Fold e.17: D-aminoacid aminotransferase-like PLP-dependent enzymes [56751] (1 superfamily)
2 domains: (1) alpha+beta: beta3-alpha2-beta2; (2) alpha/beta, a part of its mixed sheet forms barrel: n=6, S=8
Superfamily e.17.1: D-aminoacid aminotransferase-like PLP-dependent enzymes [56752] (2 families) 

Family e.17.1.1: D-aminoacid aminotransferase-like PLP-dependent enzymes [56753] (4 proteins) 
Protein D-aminoacid aminotransferase [56754] (2 species) 
Species Bacillus sp., strain YM-1 [TaxId:1409] [56755] (8 PDB entries)
domain 1: 1-120; domain 2: 121-277
Domain d1a0ga_: 1a0g A: [43276]
complexed with pmp; mutant
Details for d1a0ga_
PDB Entry: 1a0g (more details), 2 Å
PDB Description: l201a mutant of d-amino acid aminotransferase complexed with pyridoxamine-5'-phosphate
PDB Compounds: (A:) d-amino acid aminotransferaseSCOPe Domain Sequences for d1a0ga_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1a0ga_ e.17.1.1 (A:) D-aminoacid aminotransferase {Bacillus sp., strain YM-1 [TaxId: 1409]}
gytlwndqivkdeevkidkedrgyqfgdgvyevvkvyngemftvnehidrlyasaekiri
tipytkdkfhqllhelveknelntghiyfqvtrgtsprahqfpentvkpviigytkenpr
plenlekgvkatfvedirwlrcdikslnllgavlakqeahekgcyeailhrnntvtegss
snvfgikdgilythpannmiakgitrdvviacaneinmpvkeipftthealkmdelfvts
ttseitpvieidgklirdgkvgewtrklqkqfetkipkpl
SCOPe Domain Coordinates for d1a0ga_:
Click to download the PDB-style file with coordinates for d1a0ga_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1a0ga_:
- d1a0ga_ first appeared (with stable ids) in SCOP 1.55
- d1a0ga_ appears in SCOPe 2.05
- d1a0ga_ appears in SCOPe 2.07
- d1a0ga_ appears in the current release, SCOPe 2.08
 View in 3D
View in 3D