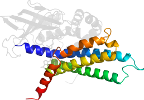
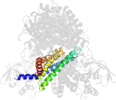
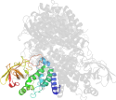
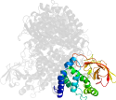Lineage for d1egdd2 (1egd D:10-241)
- Root: SCOP 1.75

Class e: Multi-domain proteins (alpha and beta) [56572] (66 folds) 
Fold e.6: Acyl-CoA dehydrogenase NM domain-like [56644] (1 superfamily)
2 domains: (1) all-alpha: 5 helices; (2) contains an open beta-sheet barrel: n*=5, S*=8; complex topology
Superfamily e.6.1: Acyl-CoA dehydrogenase NM domain-like [56645] (2 families) 
flavoprotein: binds FAD; constituent families differ in the numbers of C-terminal domains (four-helical bundles)
Family e.6.1.1: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains [56646] (9 proteins) 
Protein Medium chain acyl-CoA dehydrogenase, NM domains [56649] (3 species) 
Species Human (Homo sapiens) [TaxId:9606] [56651] (5 PDB entries)
Uniprot P11310 34-421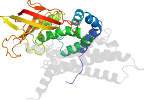
Domain d1egdd2: 1egd D:10-241 [42860]
Other proteins in same PDB: d1egda1, d1egdb1, d1egdc1, d1egdd1
complexed with fad; mutant
Details for d1egdd2
PDB Entry: 1egd (more details), 2.4 Å
PDB Description: structure of t255e, e376g mutant of human medium chain acyl-coa dehydrogenase
PDB Compounds: (D:) medium chain acyl-coa dehydrogenaseSCOP Domain Sequences for d1egdd2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1egdd2 e.6.1.1 (D:10-241) Medium chain acyl-CoA dehydrogenase, NM domains {Human (Homo sapiens) [TaxId: 9606]}
lgfsfefteqqkefqatarkfareeiipvaaeydktgeypvplirrawelglmnthipen
cgglglgtfdacliseelaygctgvqtaiegnslgqmpiiiagndqqkkkylgrmteepl
mcaycvtepgagsdvagiktkaekkgdeyiingqkmwitnggkanwyfllarsdpdpkap
ankaftgfiveadtpgiqigrkelnmgqrcsdtrgivfedvkvpkenvligd
SCOP Domain Coordinates for d1egdd2:
Click to download the PDB-style file with coordinates for d1egdd2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1egdd2:
- d1egdd2 first appeared (with stable ids) in SCOP 1.55
- d1egdd2 appears in SCOP 1.73
- d1egdd2 appears in SCOPe 2.01
- d1egdd2 appears in the current release, SCOPe 2.08

 View in 3D
View in 3D