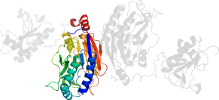
Lineage for d1emsb2 (1ems B:10-280)
- Root: SCOPe 2.07

Class d: Alpha and beta proteins (a+b) [53931] (388 folds) 
Fold d.160: Carbon-nitrogen hydrolase [56316] (1 superfamily)
4 layers: alpha/beta/beta/alpha; mixed beta sheets; contains duplication
Superfamily d.160.1: Carbon-nitrogen hydrolase [56317] (3 families) 
Pfam PF00795; some topological similarities to the metallo-dependent phosphatases and DNase I-like nucleases
Family d.160.1.1: Nitrilase [56318] (2 proteins) 
Protein NIT-FHIT fusion protein, N-terminal domain [56319] (1 species) 
Species Nematode (Caenorhabditis elegans) [TaxId:6239] [56320] (1 PDB entry) 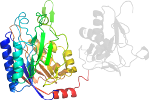
Domain d1emsb2: 1ems B:10-280 [42087]
Other proteins in same PDB: d1emsa1, d1emsb1
complexed with emc, mpd, na
Details for d1emsb2
PDB Entry: 1ems (more details), 2.8 Å
PDB Description: crystal structure of the c. elegans nitfhit protein
PDB Compounds: (B:) nit-fragile histidine triad fusion proteinSCOPe Domain Sequences for d1emsb2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1emsb2 d.160.1.1 (B:10-280) NIT-FHIT fusion protein, N-terminal domain {Nematode (Caenorhabditis elegans) [TaxId: 6239]}
matgrhfiavcqmtsdndleknfqaaknmieragekkcemvflpecfdfiglnkneqidl
amatdceymekyrelarkhniwlslgglhhkdpsdaahpwnthliidsdgvtraeynklh
lfdleipgkvrlmesefskagtemippvdtpigrlglsicydvrfpelslwnrkrgaqll
sfpsaftlntglahwetllraraienqcyvvaaaqtgahnpkrqsyghsmvvdpwgavva
qcservdmcfaeidlsyvdtlremqpvfshr
SCOPe Domain Coordinates for d1emsb2:
Click to download the PDB-style file with coordinates for d1emsb2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1emsb2:

 View in 3D
View in 3D