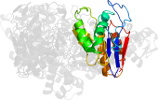
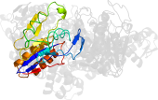
Lineage for d1g4ad_ (1g4a D:)
- Root: SCOP 1.73

Class d: Alpha and beta proteins (a+b) [53931] (334 folds) 
Fold d.153: Ntn hydrolase-like [56234] (2 superfamilies)
4 layers: alpha/beta/beta/alpha; has an unusual sheet-to-sheet packing
Superfamily d.153.1: N-terminal nucleophile aminohydrolases (Ntn hydrolases) [56235] (6 families) 
N-terminal residue provides two catalytic groups, nucleophile and proton donor
Family d.153.1.4: Proteasome subunits [56251] (3 proteins) 
Protein HslV (ClpQ) protease [56258] (3 species)
dodecameric prokaryotic homologue of proteasome
Species Escherichia coli [TaxId:562] [56259] (7 PDB entries) 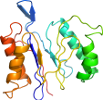
Domain d1g4ad_: 1g4a D: [41984]
Other proteins in same PDB: d1g4ae_, d1g4af_
complexed with dat
Details for d1g4ad_
PDB Entry: 1g4a (more details), 3 Å
PDB Description: crystal structures of the hslvu peptidase-atpase complex reveal an atp-dependent proteolysis mechanism
PDB Compounds: (D:) ATP-dependent protease hslvSCOP Domain Sequences for d1g4ad_:
Sequence; same for both SEQRES and ATOM records: (download)
>d1g4ad_ d.153.1.4 (D:) HslV (ClpQ) protease {Escherichia coli [TaxId: 562]}
ttivsvrrnghvviagdgqatlgntvmkgnvkkvrrlyndkviagfaggtadaftlfelf
erklemhqghlvkaavelakdwrtdrmlrkleallavadetasliitgngdvvqpendli
aigsggpyaqaaarallentelsareiaekaldiagdiciytnhfhtieelsy
SCOP Domain Coordinates for d1g4ad_:
Click to download the PDB-style file with coordinates for d1g4ad_.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1g4ad_:
- d1g4ad_ first appeared (with stable ids) in SCOP 1.55
- d1g4ad_ appears in SCOP 1.71
- d1g4ad_ appears in SCOP 1.75
- d1g4ad_ appears in the current release, SCOPe 2.08
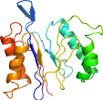
 View in 3D
View in 3D