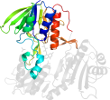
Lineage for d1nhp_3 (1nhp 322-447)
- Root: SCOP 1.71

Class d: Alpha and beta proteins (a+b) [53931] (286 folds) 
Fold d.87: CO dehydrogenase flavoprotein C-domain-like [55423] (2 superfamilies)
core: beta(3,4)-alpha(3); alpha+beta sandwich
Superfamily d.87.1: FAD/NAD-linked reductases, dimerisation (C-terminal) domain [55424] (1 family) 
both first two domains are of same beta/beta/alpha fold
Family d.87.1.1: FAD/NAD-linked reductases, dimerisation (C-terminal) domain [55425] (11 proteins) 
Protein NADH peroxidase [55432] (1 species) 
Species Enterococcus faecalis [TaxId:1351] [55433] (8 PDB entries) 
Domain d1nhp_3: 1nhp 322-447 [40196]
Other proteins in same PDB: d1nhp_1, d1nhp_2
complexed with fad, so4; mutant
Details for d1nhp_3
PDB Entry: 1nhp (more details), 2 Å
PDB Description: crystallographic analyses of nadh peroxidase cys42ala and cys42ser mutants: active site structure, mechanistic implications, and an unusual environment of arg303
SCOP Domain Sequences for d1nhp_3:
Sequence; same for both SEQRES and ATOM records: (download)
>d1nhp_3 d.87.1.1 (322-447) NADH peroxidase {Enterococcus faecalis}
gvqgssglavfdykfastginevmaqklgketkavtvvedylmdfnpdkqkawfklvydp
ettqilgaqlmskadltaninaislaiqakmtiedlayadfffqpafdkpwniintaale
avkqer
SCOP Domain Coordinates for d1nhp_3:
Click to download the PDB-style file with coordinates for d1nhp_3.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1nhp_3:
- d1nhp_3 first appeared (with stable ids) in SCOP 1.55
- d1nhp_3 appears in SCOP 1.69
- d1nhp_3 is called d1nhpa3 in SCOP 1.73
- d1nhp_3 appears in the current release, SCOPe 2.08, called d1nhpa3

 View in 3D
View in 3D