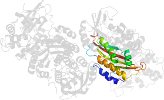
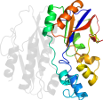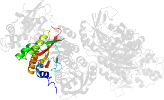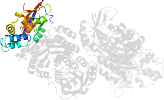Lineage for d1clid1 (1cli D:3021-3170)
- Root: SCOPe 2.08

Class d: Alpha and beta proteins (a+b) [53931] (396 folds) 
Fold d.79: Bacillus chorismate mutase-like [55297] (9 superfamilies)
core: beta-alpha-beta-alpha-beta(2); mixed beta-sheet: order: 1423, strand 4 is antiparallel to the rest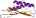
Superfamily d.79.4: PurM N-terminal domain-like [55326] (2 families) 
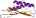
Family d.79.4.1: PurM N-terminal domain-like [55327] (7 proteins) 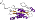
Protein Aminoimidazole ribonucleotide synthetase (PurM) N-terminal domain [55328] (3 species) 
Species Escherichia coli [TaxId:562] [55329] (1 PDB entry) 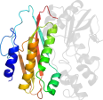
Domain d1clid1: 1cli D:3021-3170 [39819]
Other proteins in same PDB: d1clia2, d1clib2, d1clic2, d1clid2
complexed with so4
Details for d1clid1
PDB Entry: 1cli (more details), 2.5 Å
PDB Description: x-ray crystal structure of aminoimidazole ribonucleotide synthetase (purm), from the e. coli purine biosynthetic pathway, at 2.5 a resolution
PDB Compounds: (D:) protein (phosphoribosyl-aminoimidazole synthetase)SCOPe Domain Sequences for d1clid1:
Sequence; same for both SEQRES and ATOM records: (download)
>d1clid1 d.79.4.1 (D:3021-3170) Aminoimidazole ribonucleotide synthetase (PurM) N-terminal domain {Escherichia coli [TaxId: 562]}
alvgrikgvvkktrrpevmgglggfgalcalpqkyrepvlvsgtdgvgtklrlamdlkrh
dtigidlvamcvndlvvqgaeplffldyyatgkldvdtasavisgiaegclqsgcslvgg
etaempgmyhgedydvagfcvgvvekseii
SCOPe Domain Coordinates for d1clid1:
Click to download the PDB-style file with coordinates for d1clid1.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1clid1:
- d1clid1 first appeared (with stable ids) in SCOP 1.55
- d1clid1 appears in SCOPe 2.07
 View in 3D
View in 3D