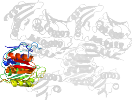
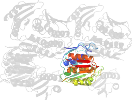
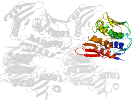
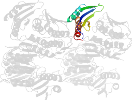Lineage for d1qmic2 (1qmi C:5-184,C:280-339)
- Root: SCOP 1.75

Class d: Alpha and beta proteins (a+b) [53931] (376 folds) 
Fold d.68: IF3-like [55199] (8 superfamilies)
beta-alpha-beta-alpha-beta(2); 2 layers; mixed sheet 1243, strand 4 is antiparallel to the rest
Superfamily d.68.2: EPT/RTPC-like [55205] (2 families) 

Family d.68.2.1: RNA 3'-terminal phosphate cyclase, RPTC [55206] (1 protein)
duplication: 3 repeats of this fold are packed together around the pseudo threefold axis
Protein RNA 3'-terminal phosphate cyclase, RPTC [55207] (1 species)
also contains an insert alpha+beta domain of the thioredoxin-like topology
Species Escherichia coli [TaxId:562] [55208] (2 PDB entries) 
Domain d1qmic2: 1qmi C:5-184,C:280-339 [39572]
Other proteins in same PDB: d1qmia1, d1qmib1, d1qmic1, d1qmid1
Details for d1qmic2
PDB Entry: 1qmi (more details), 2.8 Å
PDB Description: Crystal structure of RNA 3'-terminal phosphate cyclase, an ubiquitous enzyme with unusual topology
PDB Compounds: (C:) RNA 3'-terminal phosphate cyclaseSCOP Domain Sequences for d1qmic2:
Sequence; same for both SEQRES and ATOM records: (download)
>d1qmic2 d.68.2.1 (C:5-184,C:280-339) RNA 3'-terminal phosphate cyclase, RPTC {Escherichia coli [TaxId: 562]}
mialdgaqgegggqilrsalslsmitgqpftitsiragrakpgllrqhltavkaateicg
atvegaelgsqrllfrpgtvrggdyrfaigsagsctlvlqtvlpalwfadgpsrvevsgg
tdnpsappadfirrvlepllakigihqqttllrhgfypagggvvatevspvasfntlqlg
Xavgeyladqlvlpmalagageftvahpschlltniavverflpvrfslietdgvtrvsi
e
SCOP Domain Coordinates for d1qmic2:
Click to download the PDB-style file with coordinates for d1qmic2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d1qmic2:
- d1qmic2 first appeared (with stable ids) in SCOP 1.55
- d1qmic2 appears in SCOP 1.73
- d1qmic2 appears in SCOPe 2.01
- d1qmic2 appears in the current release, SCOPe 2.08
 View in 3D
View in 3D