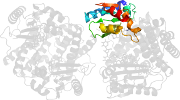Lineage for d6nmmc2 (6nmm C:126-253)
- Root: SCOPe 2.08

Class a: All alpha proteins [46456] (290 folds) 
Fold a.24: Four-helical up-and-down bundle [47161] (29 superfamilies)
core: 4 helices; bundle, closed or partly opened, left-handed twist; up-and-down
Superfamily a.24.16: Nucleotidyltransferase substrate binding subunit/domain [81593] (5 families) 

Family a.24.16.1: Kanamycin nucleotidyltransferase (KNTase), C-terminal domain [81592] (2 proteins)
N-terminal catalytic domain is followed by an all-alpha domain
automatically mapped to Pfam PF07827
Protein automated matches [379001] (3 species)
not a true protein
Species Geobacillus stearothermophilus [TaxId:1422] [379002] (4 PDB entries) 
Domain d6nmmc2: 6nmm C:126-253 [379070]
Other proteins in same PDB: d6nmma1, d6nmmb1, d6nmmc1, d6nmmd1
automated match to d1kana1
protein/DNA complex; complexed with a, mg, nmy, ppv; mutant
Details for d6nmmc2
PDB Entry: 6nmm (more details), 2.5 Å
PDB Description: ternary complex structure of the t130k mutant of ant-4 with neomycin, ampcpp and pyrophosphate
PDB Compounds: (C:) kanamycin nucleotidyltransferaseSCOPe Domain Sequences for d6nmmc2:
Sequence; same for both SEQRES and ATOM records: (download)
>d6nmmc2 a.24.16.1 (C:126-253) automated matches {Geobacillus stearothermophilus [TaxId: 1422]}
veaqkfhdaicaliveelfeyagkwrnirvqgpttflpsltvqvamagamliglhhricy
ttsasvlteavkqsdlpsgydhlcqfvmsgqlsdseklleslenfwngiqewterhgyiv
dvskripf
SCOPe Domain Coordinates for d6nmmc2:
Click to download the PDB-style file with coordinates for d6nmmc2.
(The format of our PDB-style files is described here.)
(The format of our PDB-style files is described here.)
Timeline for d6nmmc2:
- d6nmmc2 first appeared in SCOPe 2.07
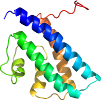
 View in 3D
View in 3D